@EricMoulton_ICM I gave it a spin. Bottom line: I think your images can be roughly aligned but for better alignment, I think you’ll need to do some fine-tuning. The result belo is not ideal but hopefully gets you started.
The images
mrinfo template_fod.mif
************************************************
Image: "template_fod.mif"
************************************************
Dimensions: 256 x 256 x 256 x 15
Voxel size: 1 x 1 x 1 x 1
Data strides: [ 1 2 3 4 ]
Format: MRtrix
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 0
0 1 0 0
0 0 1 0
mrinfo sub_fod.mif
************************************************
Image: "sub_fod.mif"
************************************************
Dimensions: 256 x 256 x 44 x 15
Voxel size: 1.0938 x 1.0938 x 3 x 1
Data strides: [ -1 2 3 4 ]
Format: MRtrix
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 -143.5
-0 1 0 -96.83
-0 0 1 -59.68
affine registration
mrregister fod_sub.mif fod_template.mif -info -type affine -mask1 mask.mif -affine affine
mrtransform sub_fod.mif -linear affine sub_fod_a.mif -template template_fod.mif
cat affine
#centre 61.8269839502482 77.71968852513824 63.5994907128613
0.988832220942192 -0.1361813446584312 -0.183502224997793 -92.14744718244954
0.04260321743823417 0.8566913214839063 -0.4320483135094917 -26.24398430748857
0.2500557102936811 0.4831384306240962 0.7718184703499078 -174.1902765935251
0 0 0 1
linear transformation
mrtransform sub_b0.mif -linear affine sub_b0_a.mif -template template_fod.mif
mrtransform sub_fod.mif -linear affine sub_fod_a.mif -template template_fod.mif
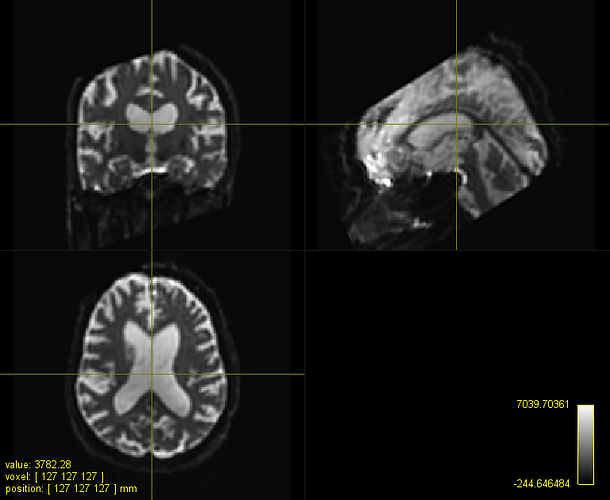
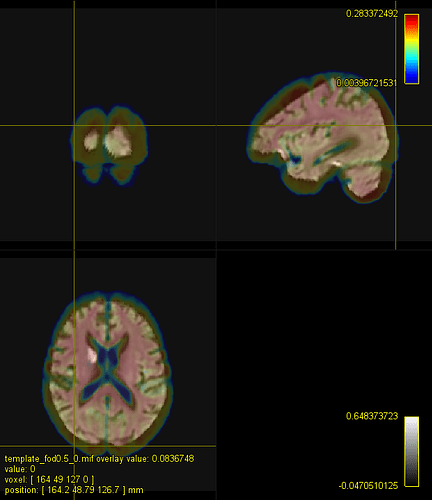
b=0 subject and template:
Phew, affine registration worked but large deformations are required… Also, there are some high intensity artefacts probably due to failure in distortion correction.
bring intensity of subject’s FOD image roughly to same range as that of the template
mrcalc sub_fod_a.mif 0.75 -mult sub_fod_as.mif
mrcalc sub_fod.mif 0.75 -mult sub_fod_s.mif
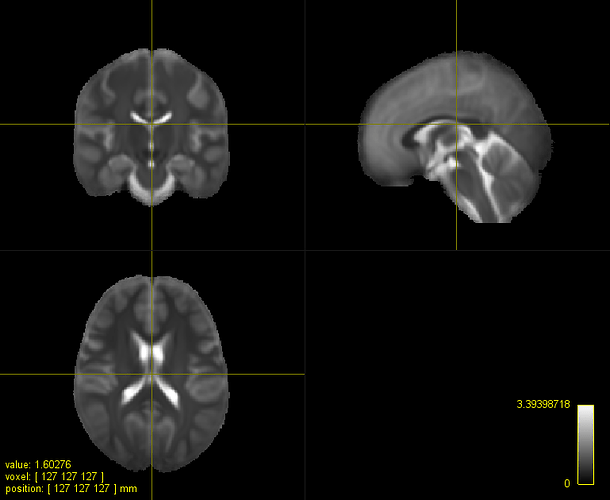
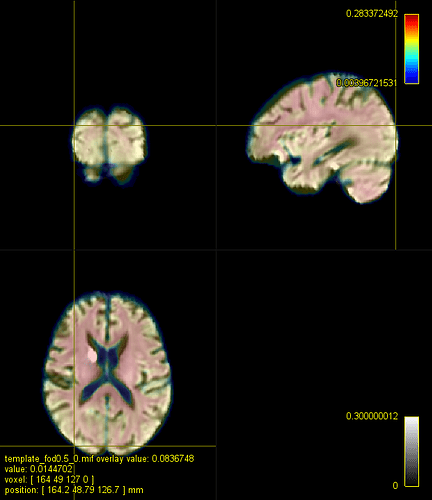
FOD subject (affine, multiplied by 0.75) and template:
nonlinear registration
We’ll need large deformations. Let’s start with lower spatial resolution than the default values but use more registration stages.
mrregister sub_fod_s.mif template_fod.mif -type nonlinear -affine_init affine -nl_scale 0.1,0.2,0.3,0.4,0.5,0.6 -info -nl_lmax 2,2,2,2,2,2 -nl_warp_full warp.mif -transformed sub_fod_asn.mif
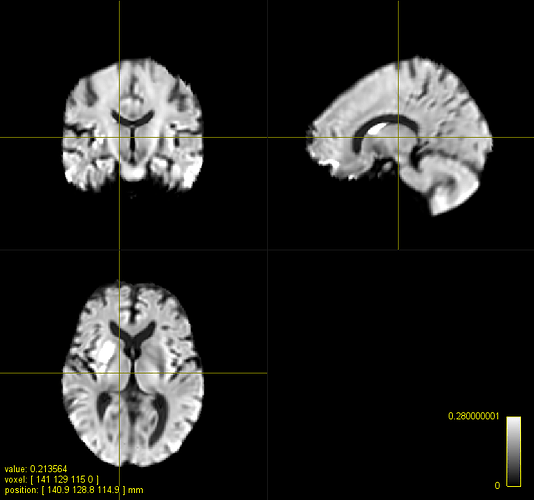
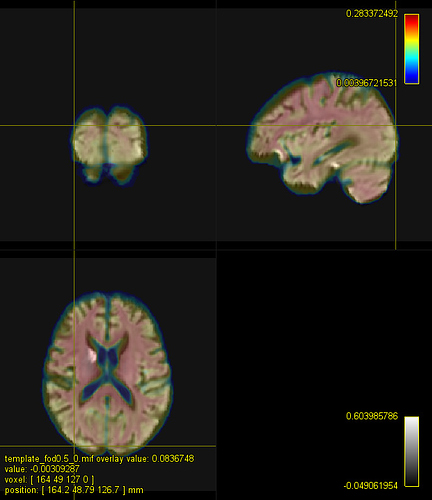
overlay is the template) without (yours) and after adjusting the intensity (sub_fod_asn.mif):
The brain size is roughly the same but the deformation is not ideal. For instance, the subject’s ventricles are not compressed enough. I’d start decreasing -nl_update_smooth -nl_disp_smooth until you get funny local distortions in your image. At that point, I think there is little we can do with the mean squared metric.
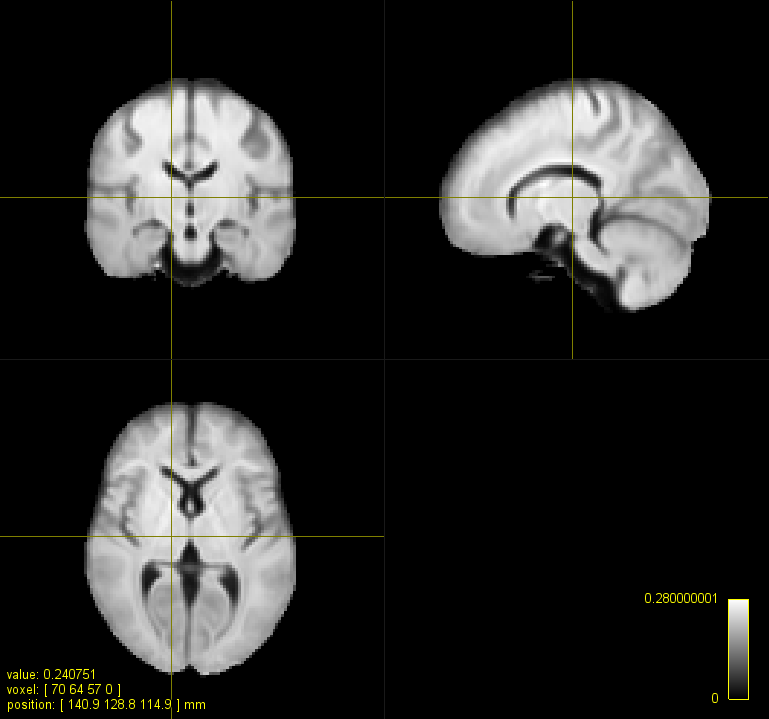
Let’s test whether the scale and lmax parameters or the intensity scaling are the cause for the better matching brain size:
mrregister sub_fod.mif template_fod.mif -type nonlinear -affine_init affine -nl_scale 0.1,0.2,0.3,0.4,0.5,0.6 -info -nl_lmax 2,2,2,2,2,2 -nl_warp_full warp_.mif -transformed sub_fod_an.mif
Worse than not scaled but better than default parameters (your image). So both.
Cheers,
Max