Hi All,
We are working with the ADNI diffusion data, and our dataset contains both single shelled and multi shelled scans.
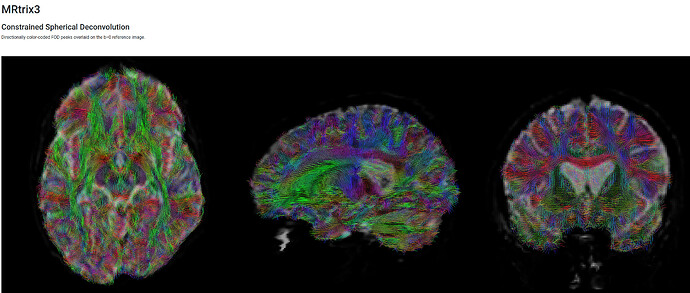
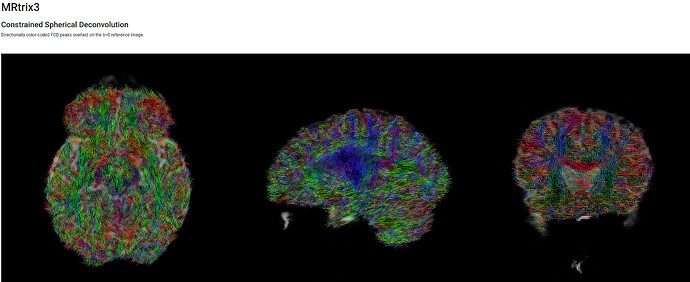
As a result, we’ve run the dhollander algorithm for the single shelled, and the msmt for the multi shelled. However, from what I can tell based on numerous forum posts, our tractography looks pretty noisy compared to some of the outputs achieved by others. Our multishelled results look better than our single shelled, but both could do with improvement.
Multi Shelled
Single Shelled
We are pretty inexperienced with diffusion data, primarily coming working with fMRI up to this point. So some basic questions:
- Is there any post processing methods we can use to refine our tractography and reduce the noise?
- Would reducing the number of tracts help?
- Is there any preprocessing steps that we’ve missed, we’ve used eddy and dwidenoise.
Any input or ideas would be massively appreciated. (I appreciate its a vague post but we’ve exhausted our admittedly limited knowledge!)
Many Thanks,
Chris
1 Like
Hi Chris,
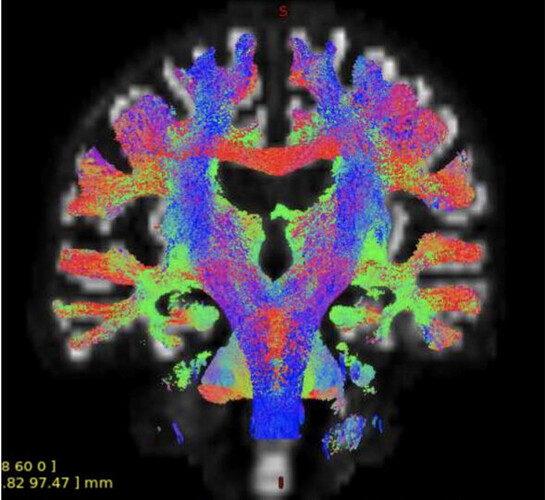
I did some experiments with ADNI2 single-shell data in the past and my tractography looked like this:
I don’t have a better picture, this is from the report we wrote at that time.
The FODs you show are expected to cover the full brain, it will be FODs in every voxel of the mask, but how the actual tractography looks like?
For the data you show, what preprocess you did? For our project we did:
- denoise (
dwidenoise)
- remove of gibbs rings artifacts (
mrdegibbs)
-
Synb0-DISCO distortion correction
- head movement, eddy current and EPI geometric distortion correction (
eddy or dwifslpreproc)
- B0 bias field inhomogeneities correction (
dwibiascorrect)
Then for the FODs we applied SS3T, but you can use MSMT with only the WM and CSF responses. Followed by mtnormalise.
Finally, we did tractography using ACT. I hope this helps.
Best regards,
Manuel
3 Likes
Hi Manuel,
Thanks for the sanity check! We followed exactly the same process, seems its just our plots that are wrong! Will plot the actual tractography.
Many Thanks,
Chris
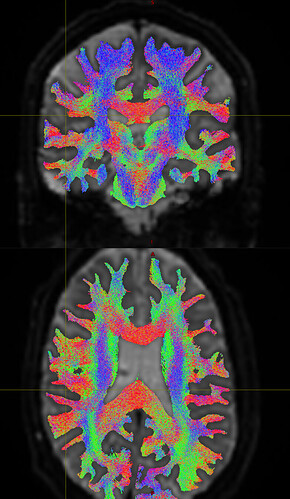
Just to check back in, this looks far more like what we are expecting! Your help is much appreciated Manuel.
@FL33TW00D those plots are FOD peaks, not streamlines. If you load the images and streamlines into mrview you should see what you’re expecting