Hi, everyone
I aim to analyze the probabilistic tractography for macaque dwi data (UNC-Wisconsin Neurodevelopment Rhesus MRI Database; single shell data) and further construct SC between ROIs. The ages of these macaque range from 2 weeks to 3 years and I want to analysis how SC changes with age.
From https://community.mrtrix.org/t/macaque-pipeline-feedback/7080, I conducted some preprocessing and further analysized of the data. However, I am not sure whether my steps are reasonable and wonder how to conduct SC analysis further, so I would like to obtain some feedback and advice.
-
preprocessing steps
(1) mrconvert -fslgrad dwi.bvec dwi.bval dwi.nii.gz dwi.mif -force
(2) mrtransform -linear UNC-Wisconsin_neurodevelopment/transform_mrtrix.txt dwi.mif dwi_trans.mif
mrtransform -strides 1,2,3,4 dwi_trans.mif dwi_trans_swap.mif
(3) dwidenoise dwi_trans_swap.mif dwi_trans_swap_den.mif
(4) mrdegibbs dwi_trans_swap_den.mif dwi_trans_swap_den_unr.mif -axes 0,2
(5) dwifslpreproc dwi_trans_swap_den_unr.mif dwi_trans_swap_den_unr_preproc.mif -rpe_none -pe_dir ap -nocleanup -debug -nthreads 8 (The diffusion data was acquired with twice refocused acquisition)
(6) dwibiascorrect ants dwi_trans_swap_den_unr_preproc.mif dwi_trans_swap_den_unr_preproc_unbiased.mif
(7) maskfilter mask.nii.gz median mask_medial.nii.gz (The mask.nii.gz is the processed mask which was provided by database)
flirt -dof 6 -ref PROCESSED/B0_LPI.nii.gz -in dwi_b0.nii.gz -omat orig2pro.txt -out b02pro.nii.gz (B0_LPI.nii.gz is the processed B0 image which was provided by database)
transformconvert orig2pro.txt dwi_b0.nii.gz PROCESSED/B0_LPI.nii.gz flirt_import orig2pro_mrtrix.txt
mrtransform -linear orig2pro_mrtrix.txt dwi_trans_swap_den_unr_preproc_unbiased.mif dwi_trans_swap_den_unr_preproc_unbiased_reg2pro.mif -template PROCESSED/B0_LPI.nii.gz (these steps aim to register my preprocessed date to the processed data which was provided by database and has high resolution)
After these preprocessing steps, I obtain the the mask and preprocessed data. But I wonder if I was right to resample dwi data before calculated response function? Additionally, are these steps reasonable and do I need to make further adjustments? -
FOD
dwi2response dhollander dwi_trans_swap_den_unr_preproc_unbiased_reg2pro.mif response_wm.txt response_gm.txt response_csf.txt -voxels 0voxels.mif -mask mask_medial.nii.gz
dwi2fod msmt_csd dwi_trans_swap_den_unr_preproc_unbiased_reg2pro.mif response_wm.txt response_wm.mif response_csf.txt response_csf.mif -mask mask_medial.nii.gz -strides 1,2,3,4 -force -info
mtnormalise response_wm.mif response_wm_norm.mif response_csf.mif response_csf_norm.mif -mask mask_medial.nii.gz -check_norm mtnormalise_norm.mif -check_mask mtnormalise_mask.mif -force -info
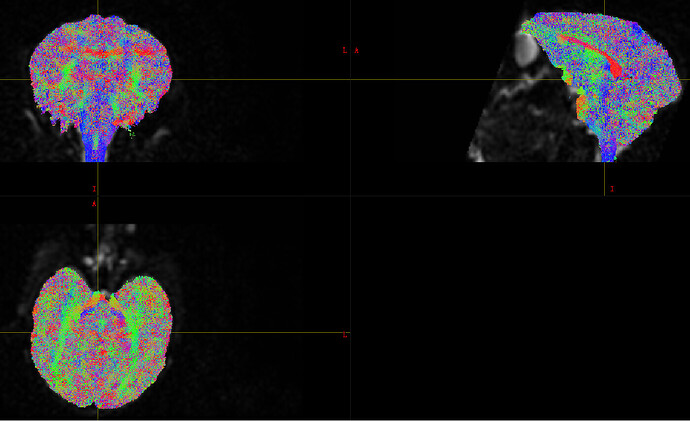
I used msmt_csd to calculate fod insteaf of ss3t_csd_beta1, as I have trouble in installing MRtrix3Tissue. Here is the wm.mif, is it looks ok? Also, I want to estimate the effect of age on SC. So do I need to average the response function (A subjuct has multiple scans)?
-
tckgen -algo iFOD2 -maxlength 250 -nthreads 8 -cutoff 0.06 -select 100M -seed_dynamic response_wm_norm.mif response_wm_norm.mif tracks_10M.tck -force -debug -info
tcksift2 -out_mu sift2_mu.txt -out_coeffs sift2_coeffs.txt -nthreads 8 tracks_10M.tck response_wm_norm.mif tracks_sift_1M.tck -force
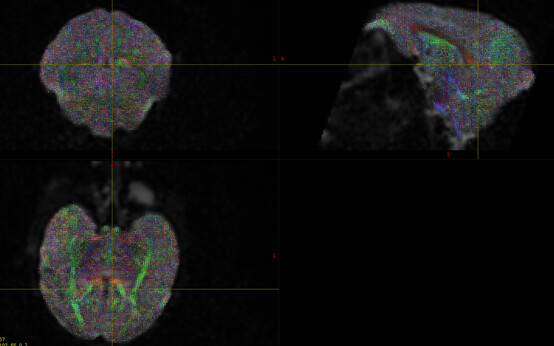
The following picture is tracks_200k.tck which was generated for ease of presentation. Is it looks right? Do I need to make my own white matter mask as seed image?
-
I have not compute SC cause I some questions about tck2connectome.
(1) Could I used sift2_mu.txt to quantify streamlines?
(2) Could I calculate SC between ROIs in template space as I also want to compute FC between ROIs with other fsfmri data?
(3)