Hello Ozzy from an Aussie! 
I’ve not had the experience of a large number of different FBA cohorts and applications coming across my desk, so I’ve not thought particularly hard about what a systematic FBA QC might look like. Certainly taking the subject FODs warped to template space (preferably with reorientation, even though that’s explicitly disabled for the sake of fixel calculations) and running an animation across them seeking gross anatomical misalignments is low-hanging fruit. Another you might want to consider is looking at, for each fixel, the number of subjects for which the value of FD is zero, i.e. no corresponding subject fixel was found. Those can influence the GLM quite strongly. Hopefully soon this work will get integrated, which will give some different options for mitigating such. Beyond that, it’s definitely something toward which community members could contribute ideas and code 
I was made aware that subject intra cranial volume is related to FC and should be corrected for, but I thought this shouldn’t be the case for FD. Could there be a correlation between FD and cranial volume as well or do you think my results are related to some sort of an error in the analysis?
So the advice you received may or may not have been downstream of this abstract. But this is actually quite an interesting point in the context of your specific work.
Over and above the experimental data shown there, the logic behind ICV influencing FC but not FD is that, within an image voxel in the middle of the WM of some small volume (e.g. 2mm x 2mm x 2mm), the fraction of the volume of just that voxel that is / can be filled with axons vs. other things possesses no logical link to the total volume of the whole head / brain. But this assumes a purely WM voxel, that’s surrounded by other WM, and WM fibre bundles that are wider in cross-section than the imaging resolution, such that differences in bundle cross-sectional area can be found morphologically using image registration. If the cross-sections of the various mesoscale WM bundles scale in appropriate proportion to total brain size, then it makes sense that the expansions / contractions necessary to align those image data to a central template space would possess some correlation with total brain size measurements. But for individual voxels within the cores of such bundles, it’s difficult to rationalize why that “packing density” / “volume fraction” would change as a function of brain size. Sure, if you were to take a brain, double its volume, but keep the same number of axons and not modify their diameters, then the voxel-wise fibre density would change along with that change in brain volume; but I simply don’t see why a healthy brain that is simply of larger volume would fail to pack in a comparable density of fibres per unit volume.
But you’re not looking at large macroscale bundles with wide cross-sections. Therein potentially lies a problem.
Imagine a WM bundle whose width & height is 1/10th the width & height of the image voxels. Firstly, this can present problems for image registration, which is what FC is derived from, but I’ll skip those details here. If there is a direct correlation between the actual physical cross-section of that bundle and brain size, this will manifest as a correlation between brain size with FD. Because within that voxel, the total volume that contains axons, and therefore contributes restriction toward a non-zero diffusion-weighted signal, increases in relation to such. So maybe in the case of looking specifically at narrow bundles, total brain size is not something you want to be ignoring.
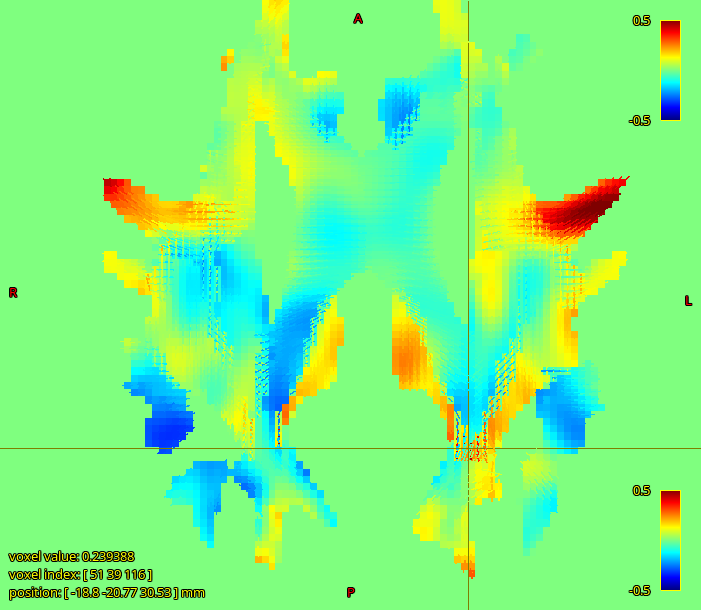
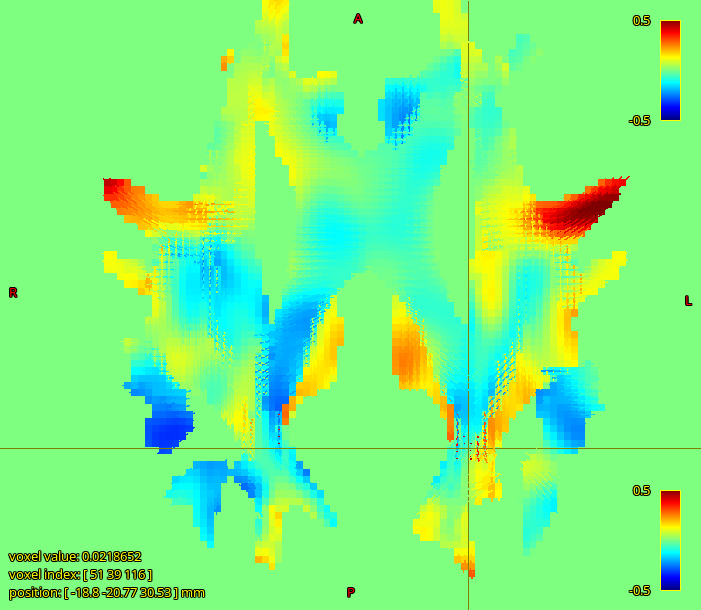
the more worrying thing I observed is that there is a strong negative correlation between our FD values and subject body size (weight and height).
Aaaaand you broke my logic  . Though this is weight and height rather than brain size, I don’t know how strongly those are correlated. Implying a link between visual system density and weight based on utilisation during exercise is probably a bit strenuous
. Though this is weight and height rather than brain size, I don’t know how strongly those are correlated. Implying a link between visual system density and weight based on utilisation during exercise is probably a bit strenuous  . Another risk is T2 effects, which is a persistently annoying confound in FD, though you’d want a reasonable argument for why it would influence your particular measurements rather than using it as an escape clause.
. Another risk is T2 effects, which is a persistently annoying confound in FD, though you’d want a reasonable argument for why it would influence your particular measurements rather than using it as an escape clause.
Also, is it a good idea to do the tractography on the FOD template or should I have done it on each FOD image and calculate a common tract for the template by warping those individual tracks?
I think that as long as you can delineate the tract on the FOD template, it makes sense to do it that way. The alternative is a lot more work, so you’d need a reasonable expectation of it being somehow beneficial to justify the effort. If the resulting fixel mask looks faithful to known anatomy and seems to be selecting quantitative data from appropriate fixels, personally I’d stick with that.
Cheers
Rob