Hi MRtrix Community,
I’m new to diffusion MRI and just started using MRtrix for fiber-tracking. I’m trying to create specific tractogram for the cingulate bundle using HCP data and not sure if I approached it in the right way. I tried the following two methods and would really appreciate any feedback or advice on my work below.
- I used the tckgen seed_image option to include a mask of cingulate and run tractogram using the following command:
tckgen -act 5TT.mif -maxlength 250 -backtrack -cutoff 0.2 -seed_image cingulate_l_combined.mif -select 1M fod.mif cingulate_l.tck
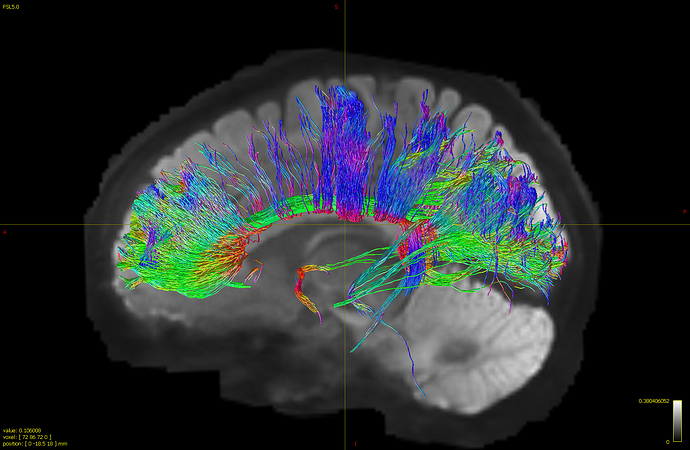
Here’s my result:
The tracts extending from cingulate look much messier that I expect…I also tried the same command using cutoff of 0.06 and the resulting tracts are even more messy as the tracts extended to the entire brain. I’m not sure if there’s a standard range of cutoff value recommend for generating tractogram? (changing cutoff value impacted my result greatly) In addition, I’m not sure how to interpret what I see, does the command I used generate all streamlines that intersect with the region of cingulate? Although cingulate is clearly shown in the tractogram, it seems to be much messier than how cingulate bundle should look like.
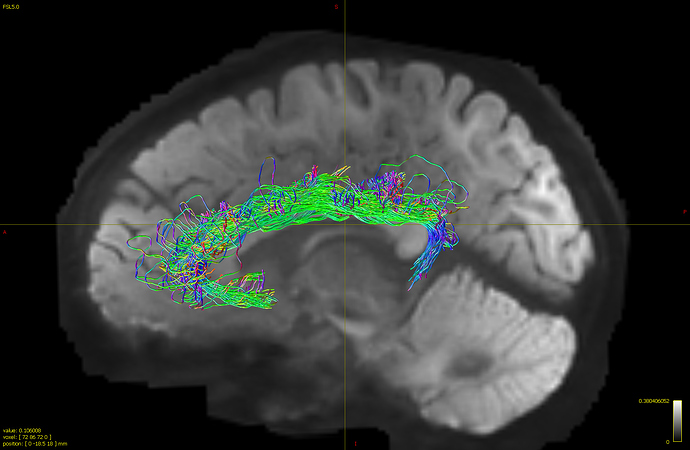
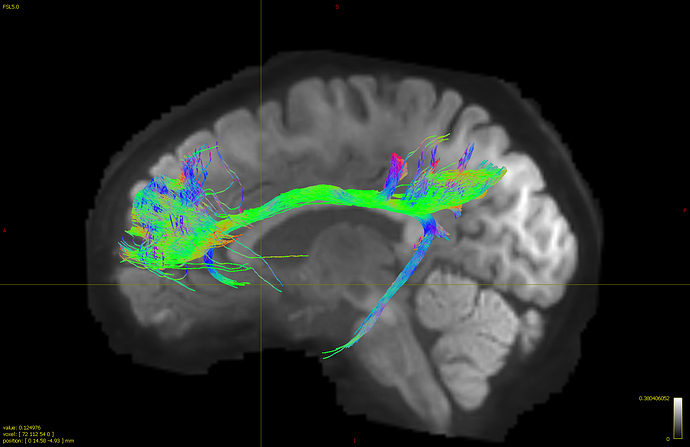
- I generated whole brain connectome first and then used tck2connectome to find tracts between four ROIs (used FreeSurfer parcellation) within cingulate (rostral anterior, caudal anterior, posterior and isthmus cingulate). After I combined the individual tractogram between each ROI using tckedit, I got the following tractogram.
tck2connectome -symmetric -zero_diagonal -scale_invnodevol whole_brain_sift.tck aparc2diff_order.mif aparc_connectome.csv -out_assignment assignments_aparc.csv
connectome2tck –nodes 2,9,21,25, –exclusive whole_brain_sift.tck assignment_aparc.csv nodes_cingulate
Although the tracts within cingulate are shown, there are some tracts that does “U-turn”, coming out from the cingulate and return back in. I’m not sure if this an accurate way of generating cingulate bundle tracts and have no idea how to clean up what look like to be unreliable tracts on the edge. Does connectome2tck generate streamlines between the two ROIs (or end points) specifically while tckgen seed_image option generate all streamlines that intersect with selected ROI, thus resulting in greater number of streamlines?
I’m not sure which approach, or if there’s other approach that would help me generate tractogram for cingulate bundle.
Any suggestion or advice is greatly appreciated and thank you so much in advance!!