I’ve not worked a whole lot with parcellations, so I don’t have a comprehensive view of what options there are unfortunately. I think the most well established and widely used is Freesurfer, it takes a very long time to run though so bear that in mind. (I’ve also heard good things about FastSurfer but I have no direct experience.)
Exactly, and you’ll find lots of other threads on this forum about coregistering T1w and DWI; FSL is a good toolbox for this task (either flirt or epi_reg, for example).
Yes indeed, a parcellation will typically consist of a label image, where the value of each voxel maps to a specific structure.
So for example, lets say you wanted an ROI for your tractography for the left primary motor cortex (left precentral gyrus). If you run Freesurfer, one of the outputs should be the parcellation file aparc.a2009s+aseg.mgz (a2009s+aseg refers to the specific atlas used). So to get just the PCG as a binary region you find the corresponding parcellation label:
11129 ctx_lh_G_precentral 60 140 180 0
and extract it with MRtrix like this:
mrcalc aparc.a2009s+aseg.mgz 11129 -eq lh_precentral_gyrus.mif
tckgen wm_fod.mif -include lh_precentral_gyrus.mif # ... etc
There is one other pretrained model (use the --tract_definition option) which is based on XTRACT segmentations, but I wouldn’t recommend it… But no harm in checking it out, it only takes 3 minutes. Other than that, you would have to retrain the model yourself with new training data, which is technically possible but not well supported.
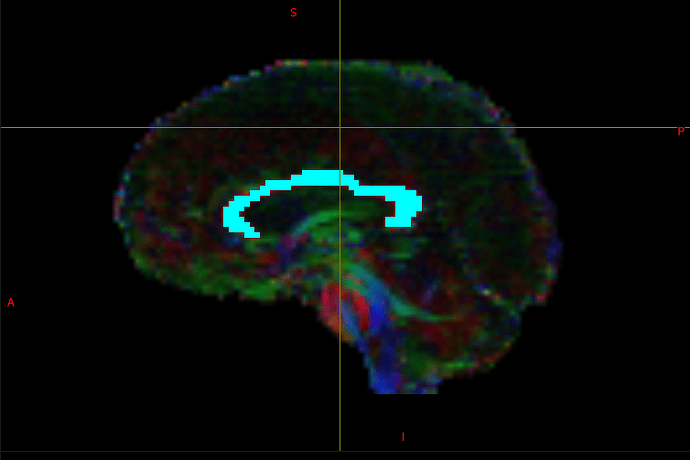
ROIs are just binary images, and TractSeg outputs binary segmentations (by default, this is the model’s inference probability thresholded at 0.5 and cleaned up a bit. You can get the raw probabilities and threshold them yourself using the --get_probabilities option).
My suggestion was that you could use the TractSeg segmentation as a tracking mask (-mask option in tckgen), since it is very big and would help clean up the image a bit. Be aware that the -mask option simply cuts off any streamlines leaving the mask, rather than discarding them. If you wanted to reject any streamline that tries to leave the mask then you’d need to invert it and use it as an -exclude ROI.