Hi,
Is there a way to register and combine together three separate images corresponding to axial, sagittal and coronal views of the same brain subject?
thank you,
Rosella
You could resample these views (volumes I assume) to the highest resolution, register (population_template, rigid) and transform and regrid (mrtransform) the original images to the common space and concatenate these (mrcat). However, a dedicated super-resolution reconstruction algorithm would higher resolution output. See related post.
Hi,
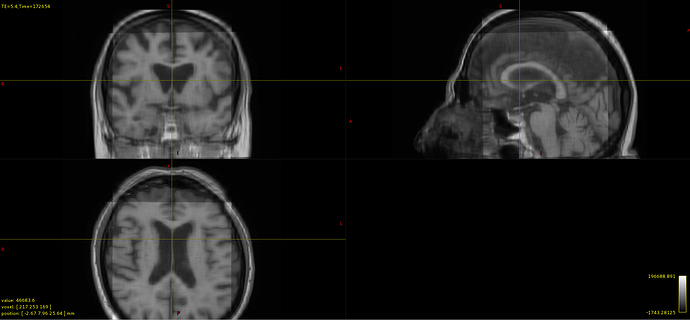
I have tried following your suggestions in order to merge and combine three views of T1 MRI images from the same subject into one volume. After applying population_template the result is the following:
I am wondering why some kind of rectangle appear on the image and if this is normal or can be avoided.
Thank you,
Rosella
Image intensity values outside the field of view are zero by default, images with partial FOV coverage transformed to the same space and averaged are therefore biasing average intensity values.
You could use population_template from the mrreg_lite2 branch, which has a -nanmask option` that takes care of this (also during registration!) or manually mark voxels outside the FOV as invalid using not-a-numbers:
For each original image i, do
mrtransform original_i.mif -template population_template.mif -nan -warp warp2template.mif warped_i.mif
then average these (nan is ignored): mrmath warped_?.mif mean new_template.mif
Sorry,
where can I find the war2template.mif if the registration mode I chose is ‘rigid’ as you suggested?
thank you
For linear transformation, replace -warp warp2template.mif with -linear trafo_i.txt. You’d get the trafo_i.txt either from the output of population_template (-linear_transformations_dir) or manually via registration of the input to the template (mrregister -type rigid using original_i.mif and population_template.mif images as input).
If the alignment is not satisfactory, make sure the images have the same intensity range (important for registration and averaging, mrcalc) and use masks in population_template even if the mask is the full image, masking in population_template will constrain registration to the intersection area of all aligned masks. Alternatively, consider using the development version of population_template (git checkout mrreg_lite2; git pull; ./build) with nan-masking and leave one out registration switched on.
I will try the development version of population_template since simply using the -nan option does not solve the interpolation problem.
Thank you
Rosella