Hi,@NicDC
Thank you for your suggestion, this really helped me a lot!! And as you recommended,
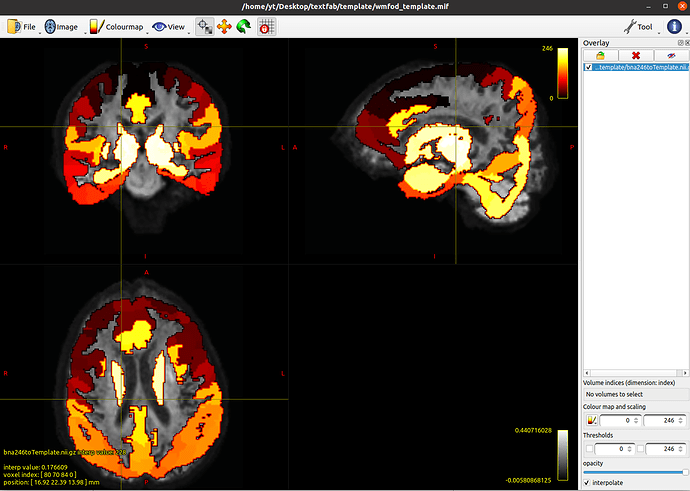
I followed this post Warping tracks files to MNI space using FSL warps moved the MNI atlas to the template successfully, but when I overlaid the Atlas2wmfod_template.nii.gz on top of my wmfod_template.mif, it looks weird. Could you please help me check my command lines?
## atlas=BN_Atlas_246_1mm.nii.gz MNI=MNI152_T1_1mm_brain.nii.gz template=wmfod_template.nii.gz
# extract the *I=0* from the wmfod_template.mif
mrconvert wmfod_template.mif wmfod_template_bad_strides.nii.gz
mrconvert wmfod_template_bad_strides.nii.gz wmfod_template.nii.gz -strides -1,2,3,4
mrconvert wmfod_template.nii.gz I0image.nii.gz -coord 3 0
# atlas to MNI152
flirt -ref ../MNI152_T1_1mm_brain.nii.gz -in ../BN_Atlas_246_1mm.nii.gz -omat bna246tomni.mat -interp nearestneighbour -dof 12
# wmfod_template to atlas
flirt -ref bna246tomni.nii.gz -in I0image.nii.gz -omat template2bna246.mat -interp nearestneighbour -dof 12
fnirt --ref=bna246tomni.nii.gz --in=I0image.nii.gz --aff=template2bna246.mat --cout=warps_template2bna246
# inverse the warp to get the atlas to wmfod_template warps
invwarp --ref=I0image.nii.gz --warp=warps_template2bna246 --out=warps_bna246totemplate
# initialise warp
warpinit bna246tomni.nii.gz inv_identity_warp_no.nii
# apply the transformation to identity warp
applywarp --ref=I0image.nii.gz --in=inv_identity_warp_no.nii --warp=warps_bna246totemplate.nii.gz --out=mrtrix_warp_bna246totemplate.nii.gz
# check the transformation
mrtransform bna246tomni.nii.gz -warp mrtrix_warp_bna246totemplate.nii.gz bna246toTemplate.nii.gz -template wmfod_template.mif -interp nearest
And in case that I misunderstand, if the registration of Atlas to wmfod_template goes well, the next step would be use the command tcktransform to transform the tracks file which generated by tckgen from the wmfod_template to the MNI Atlas, do I understand that right?
BW,
Yuting.