The mrtransform allows the spatial transformation of images when provided with a suitable linear or nonlinear transformation (“warp”). For linear transformations, MRtrix3 uses homogeneous transformation matrices stored as plain text. These can be converted from FSL and ITK-based (plain text) convention with the command transformconvert. The MRtrix3 tools expect nonlinear transformations in an image format where each voxel defines the scanner space location in the other image.
This transformation format is called a deformation field. When warping an image, these locations are used to sample intensity values in the source image space to fill the image grid in the target space. In contrast, a displacement field stores the displacements (in mm) to the other image from each voxel’s position (in scanner space). A valid MRtrix3 warp is a 4D image containing the deformation field coordinates stored as 3 volumes (x, y, z). For conversion between warp formats see warpconvert.
The following steps describe how you can use mrtransform to apply warps that have been generated from other software packages that provide registration and transformation tools not using the MRtrix3 warp format:
Warping images
The warp’s voxel grid is identical to that of the reference (“target”) image. Each spatial location in the warp specifies the offset relative to the target voxel location in scanner space coordinates from which to pull the image intensity value from the original (“source”) space image. Other software might have different file conventions such as using displacement fields and or different file and coordinate system conventions. However, you can also use mrtransform to apply warps generated from other packages that are in any format using the following steps:
First register an input/source image to a reference image using your registration package of choice. For instance, using antsRegistration on the moving image input.nii with fixed reference image reference.nii:
antsRegistration --verbose 1 --dimensionality 3 --float 0 --output [ants,antsWarped.nii.gz,antsInverseWarped.nii.gz] --interpolation Linear --use-histogram-matching 1 --winsorize-image-intensities [0.005,0.995] --transform Rigid[0.1] --metric CC[reference.nii,input.nii,1,4,Regular,0.1] --convergence [1000x500x250x100,1e-6,10] --shrink-factors 8x4x2x1 --smoothing-sigmas 3x2x1x0vox --transform Affine[0.1] --metric CC[reference.nii,input.nii,1,4,Regular,0.2] --convergence [1000x500x250x100,1e-6,10] --shrink-factors 8x4x2x1 --smoothing-sigmas 3x2x1x0vox --transform SyN[0.1,3,0] --metric CC[reference.nii,input.nii,1,4] --convergence [100x70x50x20,1e-6,10] --shrink-factors 4x2x2x1 --smoothing-sigmas 2x2x1x0vox -x [reference_mask.nii.gz,input_mask.nii.gz]
- Generate an identity (deformation field) warp using the image you wish to warp (“source”; or “moving” image):
warpinit input.mif identity_warp[].nii
- Transform this identity warp using the registration program that was used to generate the warp.
For example, if you are using the ANTs registration package:
for i in {0..2}; do
antsApplyTransforms -d 3 -e 0 -i identity_warp${i}.nii -o mrtrix_warp${i}.nii -r reference.nii -t ants1Warp.nii.gz -t ants0GenericAffine.mat --default-value 2147483647
done
- Correct the warp
The resulting 3 3D images mrtrix_warp?.nii hold the voxel-specific corresponding (x, y, z) scanner-space locations. However, locations in the target space to which no known location exists within the moving space need to be handled explicitly. Neglecting this can result in many voxels in the output mrtrix_warp pointing to the location defined by the out of bounds value, such as the origin (0.0, 0.0, 0,0) or other values as in the warp shown below (produced by the deprecated command WarpImageMultiTransform):
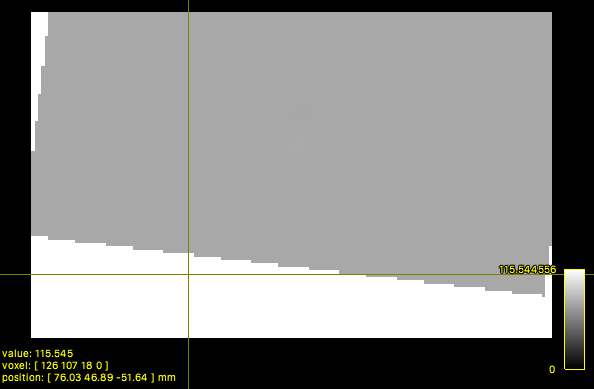
Out of bounds locations should ideally be marked by the transformation command with a unique special value that prevents using that part of the warp. MRtrix3 uses the not-a-number value (nan) for this purpose. Either specify nan as the out of bounds value if your registration package supports this or manually convert out of bound markers (here 2147483647) to vectors of nans either with the tool waprcorrect
$ warpcorrect mrtrix_warp[].nii mrtrix_warp_corrected.mif -marker 2147483647
or if that is not possible, manually for instance by transforming an empty source image (using nearest neighbour interpolation) followed by thresholding (mrthreshold) and nan-masking (see mrcalc’s -if operator).
- Warp the image
mrtransform input.nii -warp mrtrix_warp_corrected.mif warped_input_image.mif
You can check that the conversion went as expected by comparison with ANTs’ output:
mrview input_warped.mif -overlay.load antsWarped.nii.gz -overlay.opacity 0.3
Warping tracks files
To warp a tck file using a warp generated using a different software package, we need to generate an MRtrix3 compatible warp analogously to the above steps. However, warping tracks files differs from warping images due to the type of information stored: in images we pull (interpolate) intensity values sampled on the source grid to new locations on the target grid. Streamlines, on the other hand, consist of a series of vertices (coordinates defined in source space) that need to be pushed to new locations in the target space. The two operations have different spatial lookup spaces (target for images, source for tracks) and (approximately) opposing offset directions. Therefore you will need to apply the inverse transformation to warp tracks files from source to target space compared to that to warp image files.
Compared with the steps described in warping images, you will need to initialise the identity warp with respect to the reference space and amend the command used for warping the identity transform in step 2 to use the inverse warps. Note that the inverse of a series of transformations is their inverses applied in reverse order.
- Initialise warp
warpinit reference.mif inv_identity_warp[].nii
- Apply the transformation to identity warp
for i in {0..2}; do
antsApplyTransforms -d 3 -e 0 -i inv_identity_warp${i}.nii -o inv_mrtrix_warp${i}.nii -r input.nii -t [ants0GenericAffine.mat, 1] -t ants1InverseWarp.nii.gz --default-value 2147483647
done
- Fix warp
warpcorrect inv_mrtrix_warp[].nii inv_mrtrix_warp_corrected.mif -marker 2147483647
You can check that the conversion went well by transforming the reference image to the input space:
mrtransform reference.nii -warp inv_mrtrix_warp_corrected.mif reference_warped.mif
mrview reference_warped.mif -overlay.load antsInverseWarped.nii.gz -overlay.opacity 0.3
- Transform tracks file
tcktransform input.tck inv_mrtrix_warp_corrected.mif target.tck
Warping tracks files to MNI space using FSL warps
Despite the ANTs registration, the FSL warps are mostly used in the image registration, here I provide my experience to warp tracks to MNI space by using FSl warps. FSL tools might be sensitive to differences in image strides and they are defined relative to reference images. If you use pre-computed warps, please make sure (using FLS tools) that the images and warps are compatible.
**1. Generate combined warps between space of DWI,T1 and MNI **
epi_reg --pedir=-y --echospacing=2.1e-06 --epi=B0.nii.gz --t1=T1.nii.gz --t1brain=T1_brain.nii.gz --out=dwi2T1
convert_xfm -omat T12dwi.mat -inverse dwi2T1.mat
flirt -in T1_brain.nii.gz -ref B0.nii.gz -out T12dwi.nii.gz -init T12dwi.mat -applyxfm
flirt -ref MNI152_T1_3mm_brain.nii.gz -in T12dwi.nii.gz -omat T12MNI_affine.mat
fnirt --ref=MNI152_T1_3mm_brain.nii.gz --in=T12dwi.nii.gz --aff=T12MNI_affine.mat --cout=warps_T12MNI
invwarp --ref=T12dwi.nii.gz --warp=warps_T12MNI --out=warps_MNI2T1
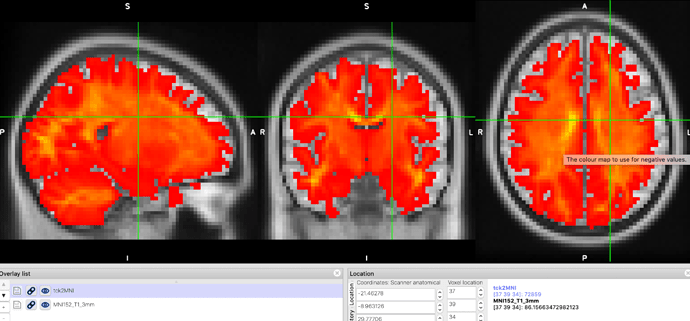
By doing so, T1 was registered to DWI space firstly; then, the transformation warp between T12DWI and MNI was generated. The transformed MNI_brain.nii is shown below:
2. Initialise warp
warpinit MNI152_T1_3mm_brain.nii.gz inv_identity_warp_no.nii
3. Apply the transformation to identity warp
applywarp --ref=b0_brain.nii.gz --in=inv_identity_warp_no.nii --warp=warps_MNI2T1.nii.gz --out=mrtrix_warp_MNI2dwi.nii.gz
Note: As far as I know, there is no option in applywarp to mark out of field of view voxels reliably. A workaround to the missing warpcorrect step is to also warp a FOV mask image and mark voxels that are not 1 in the resulting warped image as non-valid (out of field of view).
Untested example code (please confirm or fix):
mrconvert inv_identity_warp_no.nii -coord 3 0 - | mrcalc - 0 -mult 1 -add inv_identity_warp_mask.nii
applywarp --ref=b0_brain.nii.gz --in=inv_identity_warp_mask.nii --warp=warps_MNI2T1.nii.gz --out=mrtrix_warp_MNI2dwi_mask.nii.gz
mrcalc mrtrix_warp_MNI2dwi_mask.nii.gz 1 nan -if mrtrix_warp_MNI2dwi.nii.gz -mult mrtrix_warp_MNI2dwi_valid.nii.gz
Then use mrtrix_warp_MNI2dwi_valid.nii.gz instead of mrtrix_warp_MNI2dwi.nii.gz in the next steps.
4. Check the transformation
mrtransform MNI152_T1_3mm_brain.nii.gz -warp mrtrix_warp_MNI2dwi.nii.gz MNI2dwi.nii.gz
5. Transform tracks file
tcktransform ACT_50M.tck mrtrix_warp_MNI2dwi.nii.gz tck2MNI.tck -force
The red-colored image is the final transformed tracks in MNI space, and the background is MNI152_3mm.nii