Dear experts,
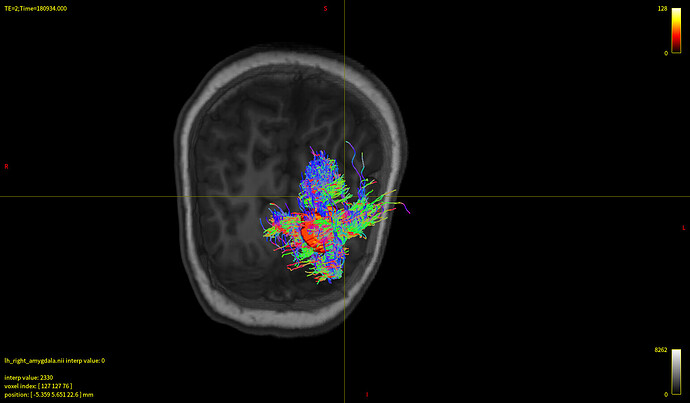
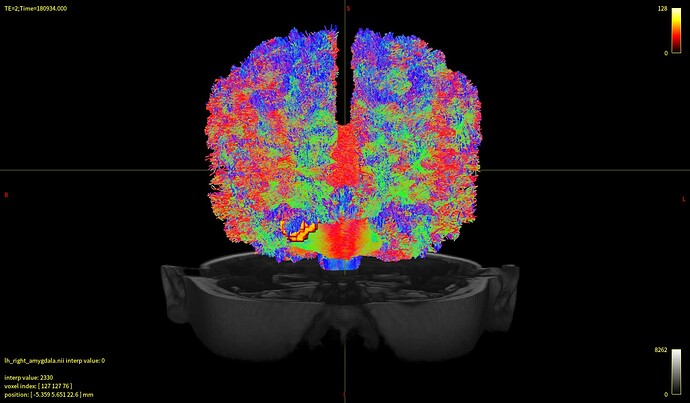
I’m sorry to trouble you. My goal is to generate tracts connecting amygdala and the whole brain. It came out that every subject’s streamlines looked almost the same as the first pic down below, while whole brain tractography looked just fine(the 2nd pic). And no matter how I modified the params(algo, angle, length, etc), the result didn’t change much. I’m wondering what’s the problem behind this.
Core commands are listed here:
-
mrthreshold -abs 0.2 ASD_1_dt_fa.mif - | mrcalc - ASD_1_dwi_mask.mif -mult ASD_1_dwi_wmMask.mif -force
-
mri_extract_label -dilate 1 aparc.a2009s+aseg.nii 54 lh_right_amygdala.nii
-
tckgen -algo iFOD2 -cutoff 0.05 -angle 45 -minlength 20 -maxlength 200 -seed_image lh_right_amygdala.nii -include ASD_1_dwi_wmMask.mif -seed_unidirectional -stop dwi_wmCsd.mif fibs_OR.tck
Thanks a lot for your kind support!
Hsu