Emre
April 18, 2024, 8:44pm
1
I have to coregister my tractography with HCP-MMP atlas. I used the codeline which is at the end.
I got this error for 2 different datasets with approximately same nodes . My suspicion is there is a problem with the files I used for coregistration.
tck2connectome: [WARNING] The following nodes do not have any streamlines assigned:
#-----------------HCP atlas prep -----------#Using files taken from this link: GitHub - mbedini/The-HCP-MMP1.0-atlas-in-FSL: A volumetric version of the HCP-MMP1 atlas conveniently imported in FSL. This repo was used in Bedini, M., Olivetti, E., Avesani, P., & Baldauf, D. (2023). Accurate localization and coactivation profiles of the frontal eye field and inferior frontal junction: an ALE and MACM fMRI meta-analysis. Brain Structure and Function, 1-21.
labelconvert MNI_Glasser_HCP_v1.0.nii HCP-Multi-Modal-Parcellation-1.0.txt ~/mrtrix3/share/mrtrix3/labelconvert/hcpmmp1_ordered.txt MNI_Glasser_HCP_nocoreg.mif
mrtransform MNI_Glasser_HCP_nocoreg.mif -linear diff2struct_mrtrix.txt -inverse -datatype uint32 MNI_Glasser_HCP_coreg.mif
Emre
April 18, 2024, 8:46pm
2
Does someone have an idea? Should I change the files which I used for coregistration
Emre
April 18, 2024, 9:31pm
4
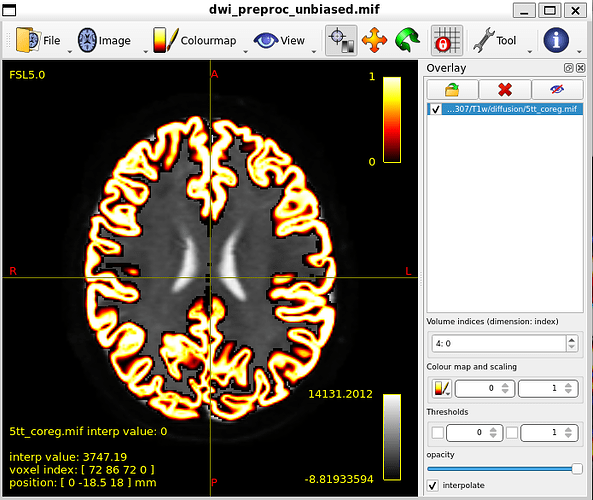
5tt_coreg.mif, overlayed
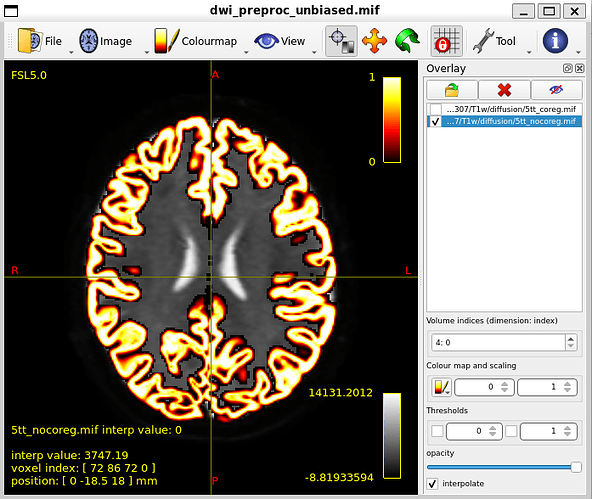
5tt_nocoreg.mif, overlayed
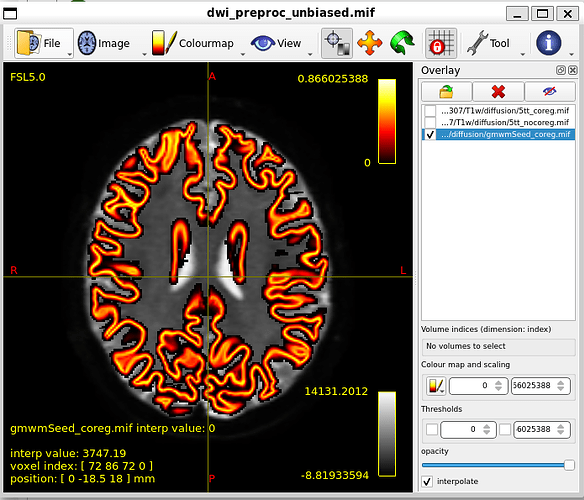
gmwmSeed_coreg.mif, overlayed
Emre
April 19, 2024, 2:11pm
5
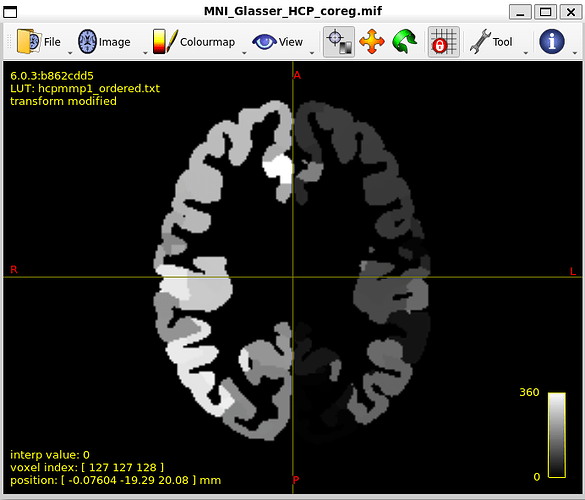
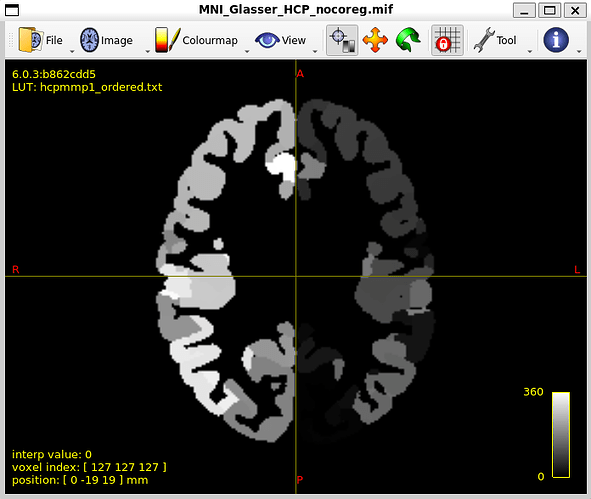
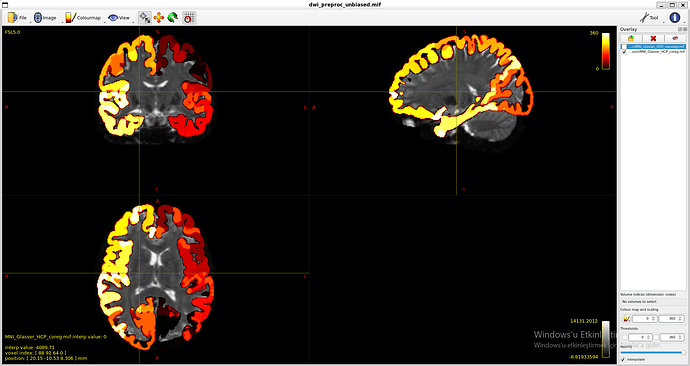
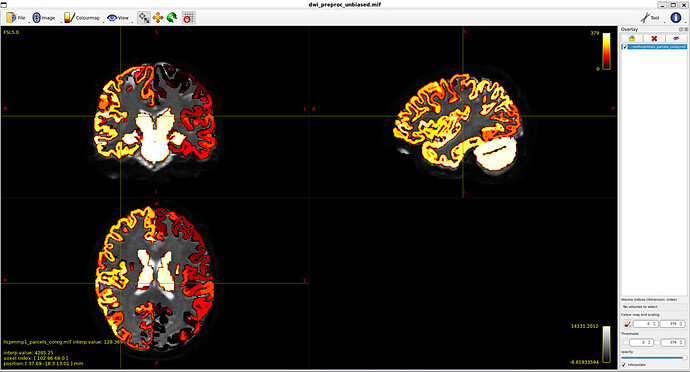
MNI_Glasser_HCP_coreg.mif is not coregistered perfectly !
I think that is the main issue with my thread. Unaligned nodes correspond to areas where the MR image does not overlap with the coreg file (such as nodes 21 and 22)
FULL CODELINE I USED
PA_DWI=$1
#---------------- preprocessing -----------------
dwi2response tournier dwi_preproc_unbiased.mif response.txt
mrgrid dwi_preproc_unbiased.mif regrid dwi_upsampled.mif -voxel 2
recon-all -i $ANAT -s $SUBJ -all
5ttgen hsvs $SUBJ 5tt_nocoreg.mif ### (NITRC: Automatic Registration Toolbox: Tool/Resource Info )
fslroi 5tt_nocoreg.nii.gz 5tt_vol0.nii.gz 0 1
flirt -in mean_b0_processed.nii.gz -ref 5tt_vol0.nii.gz -interp nearestneighbour -dof 6 -omat diff2struct_fsl.mat
5tt2gmwmi 5tt_coreg.mif gmwmSeed_coreg.mif
#----------------- Run the streamlines
#-----------------HCP atlas prep -----------#Using files taken from this link: GitHub - mbedini/The-HCP-MMP1.0-atlas-in-FSL: A volumetric version of the HCP-MMP1 atlas conveniently imported in FSL. This repo was used in Bedini, M., Olivetti, E., Avesani, P., & Baldauf, D. (2023). Accurate localization and coactivation profiles of the frontal eye field and inferior frontal junction: an ALE and MACM fMRI meta-analysis. Brain Structure and Function, 1-21.
#----------------- Constructing the connectome
Emre
April 20, 2024, 11:55pm
6
I fixed the issue by creating nocoreg and coreg filex with following codeline
mri_surf2surf --srcsubject fsaverage --trgsubject patient_recon --hemi lh --sval-annot $SUBJECTS_DIR/fsaverage/label/lh.HCP-MMP1.annot --tval $SUBJECTS_DIR/patient_recon/label/lh.HCP-MMP1.annot
mri_surf2surf --srcsubject fsaverage --trgsubject patient_recon --hemi rh --sval-annot $SUBJECTS_DIR/fsaverage/label/rh.HCP-MMP1.annot --tval $SUBJECTS_DIR/patient_recon/label/rh.HCP-MMP1.annot
mrconvert –datatype uint32 hcpmmp1.mgz hcpmmp1.mif
labelconvert hcpmmp1.mif hcpmmp1_original.txt hcpmmp1_ordered.txt hcpmmp1_parcels_nocoreg.mif
mrtransform hcpmmp1_parcels_nocoreg.mif –linear diff2struct_mrtrix.txt –inverse –datatype uint32 hcpmmp1_parcels_coreg.mif
tck2connectome -assignment_end_voxels -symmetric -zero_diagonal tracks_10M.tck hcpmmp1_parcels_coreg.mif connectome_of_patient.csv -out_assignment assignments_of_patient_sub_hcpmmp.csv
++ Now you can see how well coreg file fits