Hi,
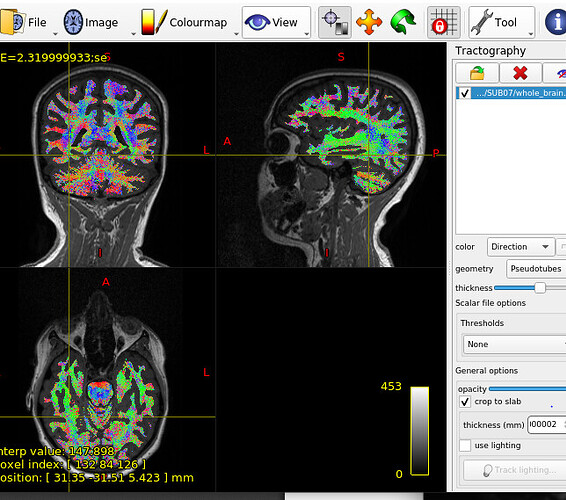
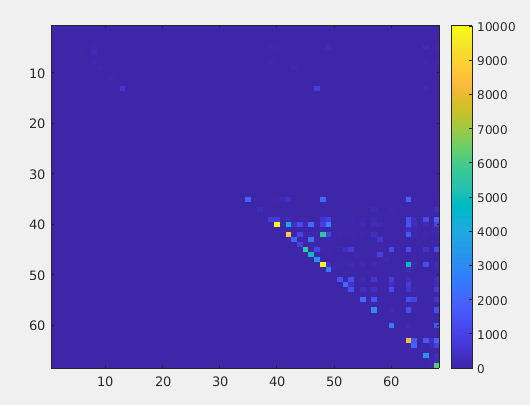
I am analyzing the DTI data collected from a local SIEMENS PRISMA scanner of healthy subjects. I preprocessed the DTI data and generated the tractogram and SC matrix using MRTrix. I did KWYK parcellation. The main issue is that the structural connectivity matrix is showing 0 values/few values/no connection for left hemisphere. Right hemisphere seems correct and showing good connections. Though tractography showed normal expected image (connections in both side) when visualized in MRTrix.
Can someone please guide me what am I doing wrong here? This is the following protocol, I am using –
cd SUBXX/
- mrconvert RAW/ep2ddiffmddw64CH90DIRAPMS.nii.gz -fslgrad bvec bval dwi_file1.mif
- mrconvert RAW/PAMS.nii.gz b0_pa.mif
- dwidenoise -force -noise noise.mif dwi_file1.mif dwi_denoised_file.mif
- mrdegibbs -force dwi_denoised_file.mif dwi_degibbs_file.mif
- dwiextract dwi_degibbs_file.mif -bzero b0_ap.mif
- mrcat b0_ap.mif b0_pa.mif b0_2.mif -axis 3
- dwifslpreproc dwi_degibbs_file.mif dwi_preproc_out.mif -rpe_pair -se_epi b0_2.mif -pe_dir ap -readout_time 0.0851 -align_seepi -eddy_options "–slm=linear " – AP and PA
or, 7. dwifslpreproc -force dwi_degibbs.mif dwi_preproc.mif -rpe_none -pe_dir ap -eddy_options "–slm=linear " – incase there is no PA - mrmath -quiet b0_ap.mif -axis 3 mean b0.nii.gz
- flirt -dof 6 -cost normmi -in t1.nii.gz -ref b0.nii.gz -omat T_fsl.txt
- transformconvert T_fsl.txt t1.nii.gz b0.nii.gz flirt_import T_T1toDWI.txt
- mrtransform -force -linear T_T1toDWI.txt RAW/t1.nii.gz aligned_t1.nii.gz
- mrconvert aligned_t1.nii.gz aligned_t1.mif
- 5ttgen -force -quiet fsl aligned_t1.mif 5TT.mif
- 5tt2gmwmi -force -quiet 5TT.mif gmwmi.mif
- dwi2response msmt_5tt -force dwi_preproc.mif 5TT.mif wm.txt gm.txt csf.txt
16… dwi2fod -force -quiet msmt_csd dwi_preproc.mif wm.txt WM_FODs.mif – multishell data - tckgen -force -quiet -seed_image gmwmi.mif -act 5TT.mif -backtrack -crop_at_gmwmi WM_FODs.mif whole_brain.tck -select 1000000
- Parcellation image
- tcksift whole_brain.tck WM_FODs.mif wholebrain_sift.tck
- tck2connectome -force whole_brain.tck parcels_DK34.nii SC_matrix_numfib.csv
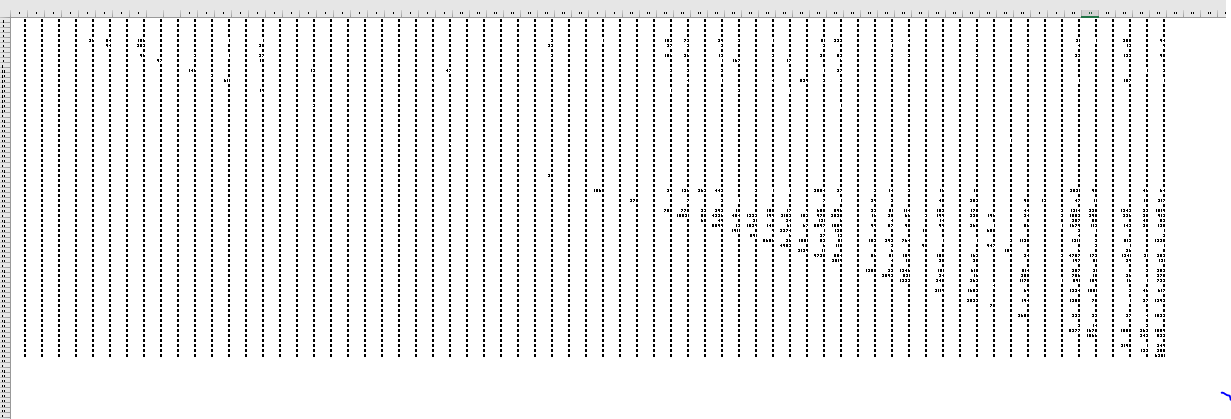
The data looks like this -
Structural connectivity matrix
Tractography screenshot -
Thanks and regards,
Swagata