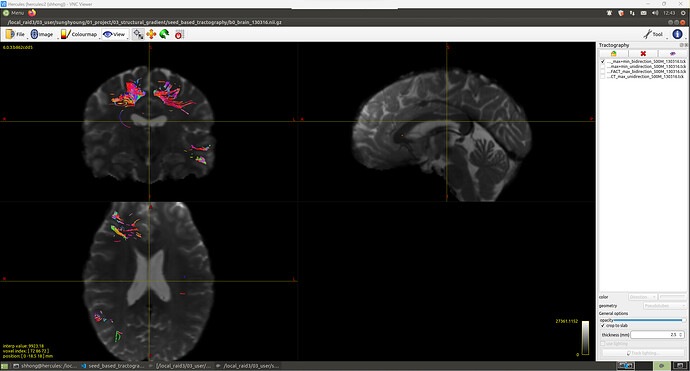
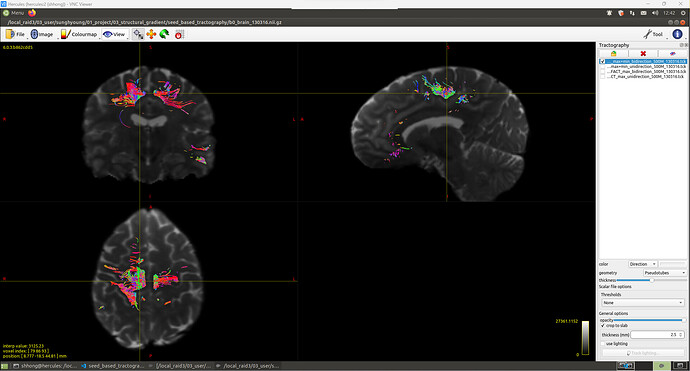
Hello, MRtrix3 experts!
I’m trying to do seed-based tractography to 1) get all the streamlines from one roi to all the other brain voxels, 2) get all the streamlines from one roi to the other.
But the results were different with what I expected. Streamlines did not spread throughout the brain, but only in local areas.
I ran the following commands. Algorithm which I used is FACT.
- getting all the streamlines from one roi to all the other brain voxels
os.system(f'tckgen {data_dir}/dhollander_FOD_wm_norm_{sub}.mif {output_dir}/ctx_com_{algorithm}_max_unidirection_500M_{sub}.tck \
-act 5TT2b0_{sub}.mif -crop_at_gmwmi -maxlength 250 -algorithm {algorithm} -seeds 50M -seed_unidirectional \
-seed_image {output_dir}/vos_ctx_com_130316_FA_max_top5.nii.gz')
os.system(f'tckgen {data_dir}/dhollander_FOD_wm_norm_{sub}.mif {output_dir}/ctx_com_{algorithm}_max_bidirection_500M_{sub}.tck \
-act 5TT2b0_{sub}.mif -crop_at_gmwmi -maxlength 250 -algorithm {algorithm} -seeds 50M \
-seed_image {output_dir}/vos_ctx_com_130316_FA_max_top5.nii.gz')
- getting all the streamlines from one roi to the other
os.system(f'tckgen {data_dir}/dhollander_FOD_wm_norm_{sub}.mif {output_dir}/ctx_com_{algorithm}_max+min_unidirection_500M_{sub}.tck \
-act 5TT2b0_{sub}.mif -crop_at_gmwmi -maxlength 250 -algorithm {algorithm} -seeds 50M -seed_unidirectional \
-seed_image {output_dir}/vos_ctx_com_130316_FA_max_top5.nii.gz -seed_image {output_dir}/vos_ctx_com_130316_FA_min_top5.nii.gz')
os.system(f'tckgen {data_dir}/dhollander_FOD_wm_norm_{sub}.mif {output_dir}/ctx_com_{algorithm}_max+min_bidirection_500M_{sub}.tck \
-act 5TT2b0_{sub}.mif -crop_at_gmwmi -maxlength 250 -algorithm {algorithm} -seeds 50M \
-seed_image {output_dir}/vos_ctx_com_130316_FA_max_top5.nii.gz -seed_image {output_dir}/vos_ctx_com_130316_FA_min_top5.nii.gz')
The other results look similar.
Can you tell me what I did wrong and what I should do to do the work I wanted?
Any response will help me!
Joseph