Hello Community:
I am working with humanbrain set of DTIs
the preprocessing does not seem to help with the AP PA distorsion
as you can see in the white mater FOD image .
I am using this command for the preproc: dwifslpreproc all_dti.mif all_dti_preproc.mif -nocleanup -pe_dir AP -rpe_pair -se_epi b0_pair.mif -eddy_options " --slm=linear --data_is_shelled"
anyone knows how to denoise it differently for this specific problem?
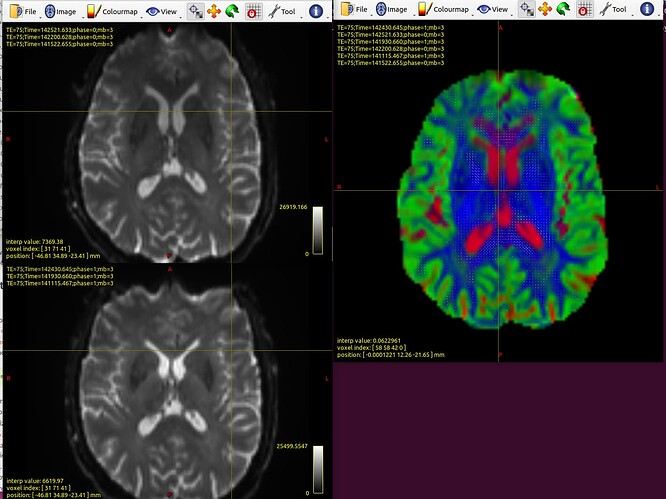
left images are my B0pair
left up PA B0mean
left down AP B0 mean
right: dedoubled frontal horns in my FODs image
thank you
z