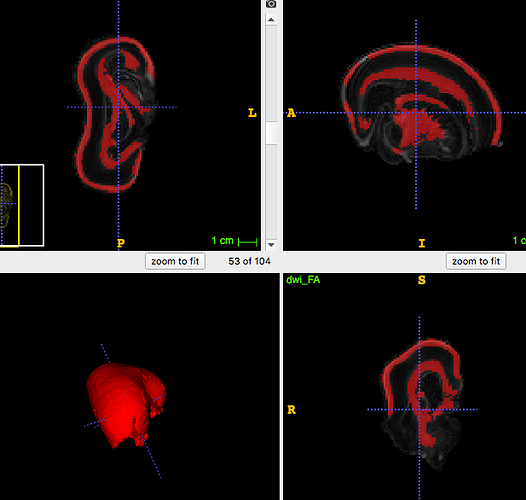
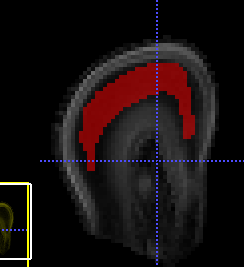
Also when running the dwi2fod spherical deconv command, do I enter the whole brain mask, or the mask attached in this picture.
So you are suggesting I have 2 masks, instead of one? how would the dwi2fod command go??
The mask used for the dwi2fod command specifies which voxels the CSD algorithm is applied to, and which voxels will contain a zero FOD. So this should be a mask of the entire brain, regardless of any masks that may be subsequently used for tractography. If you were to use these masks as the input to dwi2fod, it would be impossible to track between the two areas, since there would be no fibre orientation estimates in the space between them.
So I want to use this mask as the starting and end point of my tractography.
This is slightly difficult to apply, either using seed / include regions in tckgen or include regions in tckedit. For instance, consider the former (tckgen) case, where this ROI is provided both as the seed image and an inclusion mask. When a streamline seed point is determined, the algorithm immediately checks this point against any include regions, and determines straight away that the streamline has ‘traversed’ the include region. Therefore the streamline would then be free to propagate anywhere in the brain, and would be ‘accepted’ and written to the output file as it has successfully traversed all include regions.
It is more typical in targeted tracking experiments to use one ROI as the seed region, and another ROI as an include region, thereby requiring that streamlines must traverse from the first region into the second.
… and the -include option should be the whole brain mask for the tckgen command
This would have no effect. Include regions (either in tckgen or tckedit) are used simply as filter mechanisms, to remove any streamline that fails to traverse all include regions. If you provided a whole-brain mask as an include region, it is (just about) guaranteed that all streamlines will possess at least one point that lies within this mask, and therefore such an include region would not perform any selective inclusion / removal of streamlines. It is more typical to provide a whole-brain mask image to tckgen using the -mask option, which terminates any streamline that attempts to propagate outside the ROI.
I just wan to run some tractography from the cortex to the inner ventricle thingy
I think this is probably the clearest indication of the experiment you’re trying to do. Since you want to track from the cortex, your seed ROI should contain cortex only. You want all tracks to go to ‘the inner ventricle thingy’: therefore only this region should be provided as an include ROI, so that any track that fails to traverse it will not be included in the output track file. But remember: If your include region also includes cortex, then any streamline seeded in the cortex would immediately be determined to have successfully reached this include region, and therefore this constraint would not be applied as you had intended.
a) Yeh… like you were saying, the gradients, is there a really easy way to spot what dimension needs to be fixed?
If you are interested, you can try checking out the dwigradcheck branch in Git, and running the dwigradcheck script on your DWI. It’s still in development, but the goal is to provide an automated way of estimating errors in the diffusion gradient table, and provide the appropriately corrected table if requested.