Dear experts,
I am getting a very strange result when trying to normalise my tracts to MNI space (see below). Overall, the normalisation seems to be working but the inferior most vertices of the corticospinal fibers connect to what looks like the midpoint of the image. I have tried to use two different versions of MRTrix (0.3.15-32-g01c8f960 and 3.0_RC1-189-gbe66a20a).
I am using HCP data and these are the commands that I have used:
warpinit MNI_T1w.nii.gz no-warp.nii
applywarp -i no-warp.nii -r Native_T1w -w standard2acpc_dc.nii.gz -o warp.nii.gz
gunzip warp.nii.gz
mrconvert warp.nii -datatype float32 -stride -2,3,4,1 warp.mif
tcknormalise native_tract.tck warp.mif mni_tract.tck
I have also tried doing it without the mrconvert step… same problem
The native tract looks like this:
The warp looks like this
Does anyone have any idea what might be going wrong?
Not sure what is going on but I guess that some points are out of FOV and get mapped to [0,0,0]. I’d check warp.nii.gz for extreme values.
By the way, MRtrix3 supports .nii.gz natively and you don’t need to change the voxel stride before tcknormalise.
Thanks for the response max, that’s what it seems to be doing but it doesn’t map them to [0,0,0] exactly… which is even more odd.
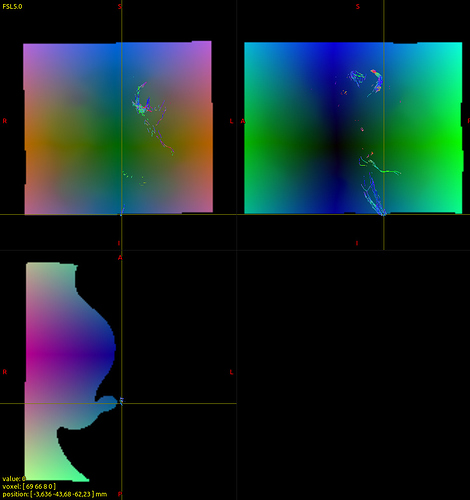
I’m checking warp.nii.gz in a viewer and I think I know what may have happened. The streamlines at the end of the corticospinal tract seem to be in an area that is outside the warped FOV and contains zero values (the reason they don’t map exactly to [0 0 0] I’m still unsure about but perhaps some interpolation is involved. In any case here are the images:
As a workaround I created a tracking mask that ensures that the tracking does not proceed to the bottom of the brainstem… this solved the problem (though it’s not the most elegant solution).
As a side note I have been checking the strides of the no-warp image (obtained from the warpinit command:
warpinit MNI_T1w.nii.gz no-warp.nii
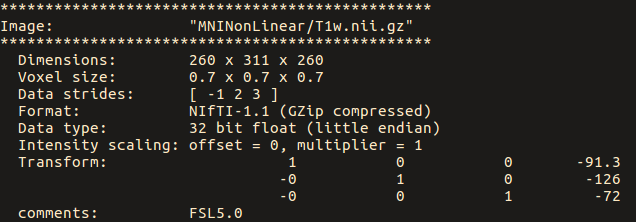
While the MNI T1 image has strides of [-1 2 3]:
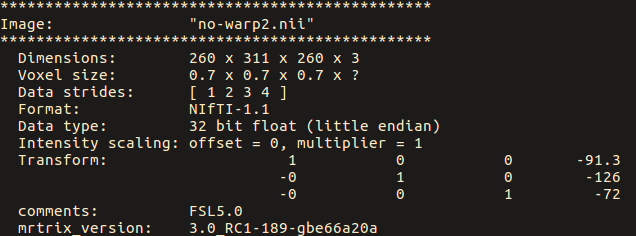
the no-warp.nii has strides of [1 2 3 4]
Should this be of concern due to flipping of the 1st dimension?
You can correct for this using warpcorrect.
The strides are handled consistently within MRtrix3 so there should be no problem. However, you might need to change the strides if you want to use the warp in other tools, for instance, FSL.
1 Like
Thanks max! looks like this is exactly what I was after.