Dear MRtrix community,
I have some questions about the streamline length values, both before and after applying the SIFT algorithm.
I generated the whole-brain tractographies of some HCP subjects following the guidelines of the ISMRM tutorial (http://mrtrix.readthedocs.io/en/latest/quantitative_structural_connectivity/ismrm_hcp_tutorial.html), except from the total initial number of streamlines that I fixed to 5M. I used 3 different algorithms: CSD muti-shell, CSD single-shell (b=1000) and DTI single-shell (b=1000).
I have two issues:
-
I haven’t set a value for the minimum length of the streamlines, thus accepting the default value that when using ACT is equal to 2 times the voxel size (i.e. 2 x 1.25 = 2.5 mm).
However, when I calculate the minimum length of the streamlines using tckstats, I get a value close to 2.1 mm for each subject when using CSD, and close to 1.4 when using DTI. How is it possible? -
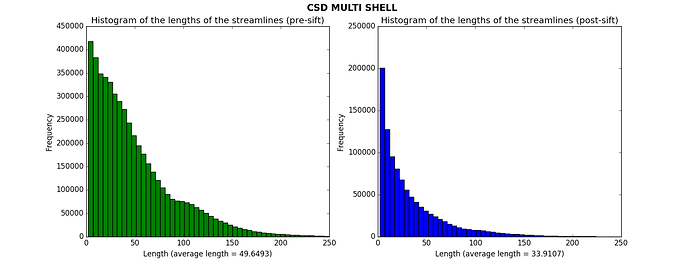
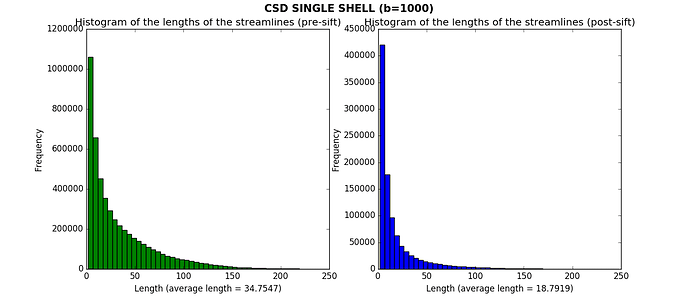
After CSD, I then applied the SIFT algorithm to have 1M streamlines. I calculated the histograms of the lengths of the streamlines before and after applying SIFT. Here what I obtained for one subject (for the other subjects the results are similar).
Is it plausible that I got such a large amount of short streamlines? It seems to me that the average lengths are pretty low, especially after having applied SIFT.
Any suggestion would be highly appreciated.
Thanks a lot,
best,
Giulia