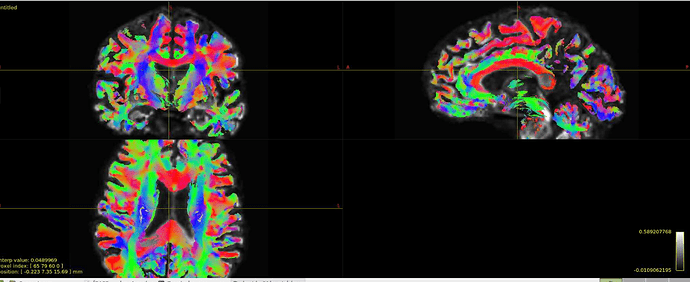
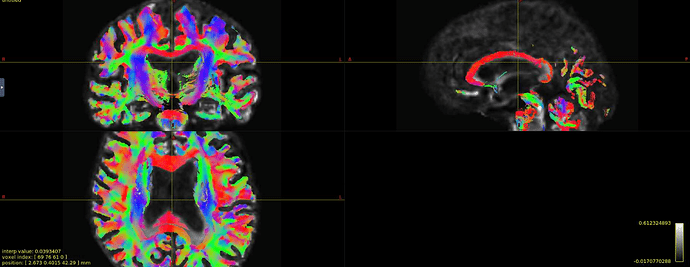
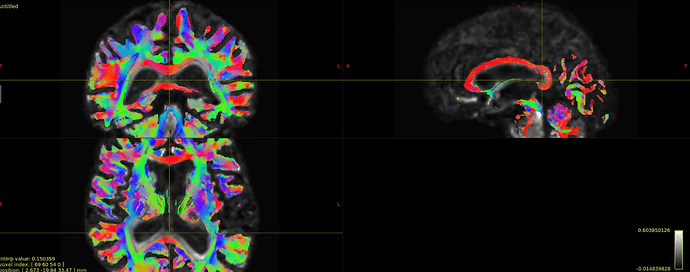
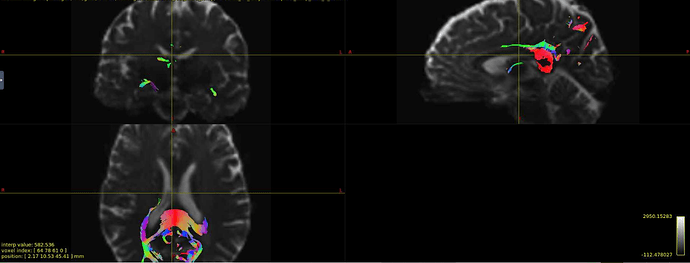
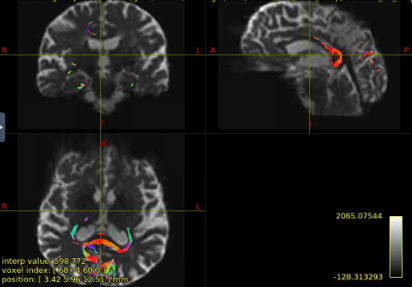
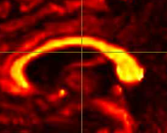
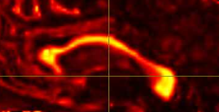
Hi all! I’m comparing network parameters using CSD vs DTI tractography algorithms for part of my dissertation work. The tractography for CSD works well, but I’m having a slight issue for some of the DTI cases. It appears that streamines are normal throughout most of the corpus callosum, but in some cases, the streamlines in the posterior corpus callosum are completely missing. This is my command:
tckgen dwi.mif tractography.tck -seed_gmwmi seed_gmwmi.mif -act 5ttgen.mif -select 50M -algorithm Tensor_Prob -force
All images in the command above are in template space. I’ve tried using -seed_dymamic and -seed_image instead of -seed_gmwmi, lowering the -cutoff value, and using -Tensor_Det instead of -Tensor_Prob. All of these choices give me similar looking output.
Any help with how to fix this would be greatly appreciated! Is this even an issue? Thank you!
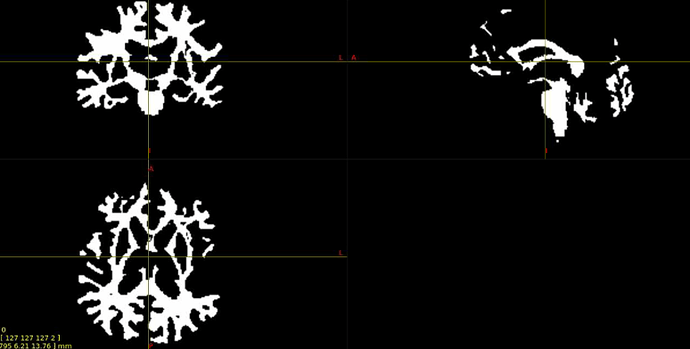
Interesting… From the coronal slices it seems like there are other WM regions devoid of tracks, such as the uncinate fasciculus. The missing part of the corpus callosums suggests that the tapetum is missing, too, which also goes to the temporal lobe. There may be something off in the diffusion data in the temporal lobes. What happens if you seed the missing region in the corpus callosum?
Thanks for your response! If I seed from the missing regions, I still see a similar pattern. I’ve also tried changing the -angle option from the default to 70 and 80, and the -step option to .2 and .08, and the same pattern persists. Since my CSD data looks normal, I’m thinking that maybe the tensor algorithm is undertracking certain regions.
It’s hard to tell from the small images, but it seem counterintuitive that a seed region would have zero streamlines going through it… unless the underlying data does not support a streamline. Can you post an image of your FA map and zoom in the callosum? Maybe there is a big hole of FA values in that region.
Yes, I can post the FA maps! I also thought that it was weird that there were no streamlines from the corpus callosum, but the FA maps look okay.
Have you checked your 5tt images? Perhaps the posterior corpus callosum is not classified as white matter on those.
1 Like
Hi! No, the 5tt images look fine as well.
When I run CSD tractography, the images look good; the problem only occurs with DTI tractography. This is leading me to think that the problem is with my dwi image, as that’s the only thing (besides the algorithm) that is changing from the FOD image. I’m running my analyses in template space. Because my template and dwi image have different dimensions and need to stay 4D, I’ve had to get creative with how to register my dwi image to the FODs in template space. Here are two ways that I’ve tried:
1.#Use mrregister to register the FOD in template space
mrregister ID_FOD.nii.gz ID_wmfod_intra_template.nii.gz -nl_warp ID_subject2template_warp_FOD_8_87.mif ID_template2subject_warp_FOD_8_27.mif -transformed ID_FOD2temp.mif -force
#Use mrtransform to take the warp from the command above to transform the dwi image to the FOD2templace space
mrtransform ID_DWI_DN_DG_ECC.mif -warp ID_subject2template_warp_FOD_8_87.mif ID_dwi2fodtemp.mif -force
2.#Run flirt to register the dwi to the FOD in template space:
flirt -dof 12 -in ID_DWI_DN_DG_ECC.nii.gz -ref ${id}_FOD2temp.nii.gz -out ID_DWI_DN_DG_ECC_flirt_2FOD2temp.nii.giz -omat ID_DWI_DN_DG_ECC_flirt_2FOD2temp.mat
#transformconvert to convert the transformation matrix for mrtrix
transformconvert ID_DWI_DN_DG_ECC_flirt_2FOD2temp.mat ID_DWI_DN_DG_ECC.nii.gz ID_FOD2temp.nii.gz flirt_import ID_DWI_DN_DG_ECC_2FOD2temp_transform.txt -force
#mrtransform to transform the dwi to FOD in template space
mrtransform ID_DWI_DN_DG_ECC.mif -linear ID_DWI_DN_DG_ECC_2FOD2temp_transform.txt -template ID_FOD2temp.mif ID_DWI_DN_DG_ECC2FOD2temp.mif -force
The ID_dwi2fodtemp.mif images and the ID_DWI_DN_DG_ECC2FOD2temp.mif images are the dwi images that I’m putting into my tckgen command. I’ve looked at the images in mrview, and the registration to the template space looks good. Is there something wrong with the way that I’m registering the dwi images to the FOD2template images that could be messing with tckgen, or does the Tensor_Prob algorithm just undertracking certain regions in the brain?
It seems like you did warp your DWIs to template space, but did not modify your bvecs accordingly. This would give you incorrect estimations of the direction of the diffusion tensors. FA maps may look OK, but the principal diffusion direction could be all wrong and induce sharp-turning tracks that get truncated. I’m not so sure if this also applies to the transformation of FODs, you may want to check that things point in the right direction.
Doing DWI processing in native space is always much easier. You can then transform your derived scalar metrics to standard space without worrying about your bvecs.
Got it! I assumed that since my files were in .mif format that the bvecs would be modified automatically, but it seems as if mrconvert only transforms the image data. If I did want to process the dwi image in template space, what would be the best way to modify the bvecs?
1 Like
You may be correct that the bvecs were transformed inside the mif file, it is very likely that mrtransform is smart enough to do that. If that is the case, mrinfo on the native and std-space versions of your dwis would differ. Can you please confirm?
Unfortunately, mrinfo shows that the bvecs are still the same. Is there any MRtrix or FSL code I can run to rotate the bvecs please?
Thank you to everyone for your help with this issue! I have decided to rerun my images in native space. This should take care of the corpus callosum problem and allow streamlines to reach throughout the whole brain without having to worry about the bvecs.