Hello,
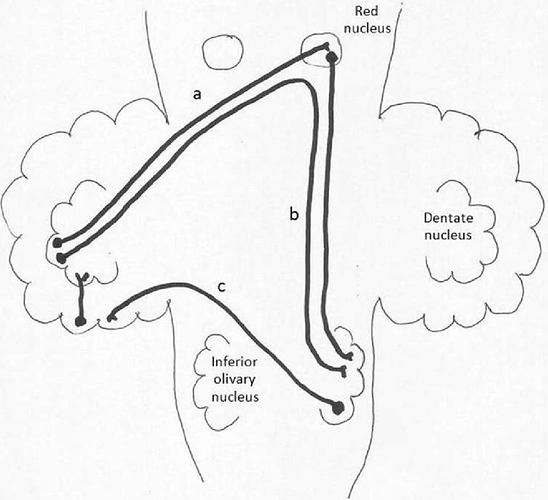
I’m working on a 7 Tesla 32 directions DTI acquisition, specifically I’m trying to reconstruct the fibers of the dentato-olivary pathway. These fibers originate from the dentate nucleus, pass through the omolateral superior cerebellar peduncle and reach the contralateral red nucleus: at this point a proportion of them synapses with red nucleus neurons, from these post-synaptic neurons a new axon originate and directs downward to reach the omolateral olive; another proportion of fibers come close to the red nucleus, but does not synapse, instead it turns downwards directly. All these fibers are part of the same fasciculus and can not be distinguished using anatomical criteria.
I have generated the 5tt segmentation for the tckgen command, and manually edited it to improve it. All the structures mentioned above are considered white matter.
From the results obtained I was able to extract two bundles of fibers: one going from the dentate nucleus to the contralateral red nucleus and terminating there, and another going from the dentate nucleus, reaching the contralateral red nucleus and continuing downward to the olive. This would be theoretically consistent with the anatomical reality mentioned above.
My question is: can MRtrix, in specific cases such as this one, actually distinguish between synapting fibers and non-synapting ones?
The possible explanation I’ve come up with is that synapting fibers have a rapid change of orientation (I imagine them as two sides of a triangle) and thus could produce, at the synapse point, an ODF with two distinct double lobes (one for the incoming fibers and the other for the emerging fibers). On the other hand non-synapting fibers reasonably have a more progressive change in orientation (with a parabolic-like shape) and thus could produce a single, wider double lobe in the ODF in the synapse point.
I assumed that in a single voxel there are only fibers of the same type (i.e. synapting or non-synapting).
Is this scenario realistic? If this was the case, would tckgen terminate fiber with a rapid change in direction, while continuing those with a progressive change in direction?
Is there a way I can extract data that can help me investigate if my supposition is realistic or not (such as the ODF in the presunt synapse points)?
This is a scheme of the dentato-olivary pathway:
Thank You.