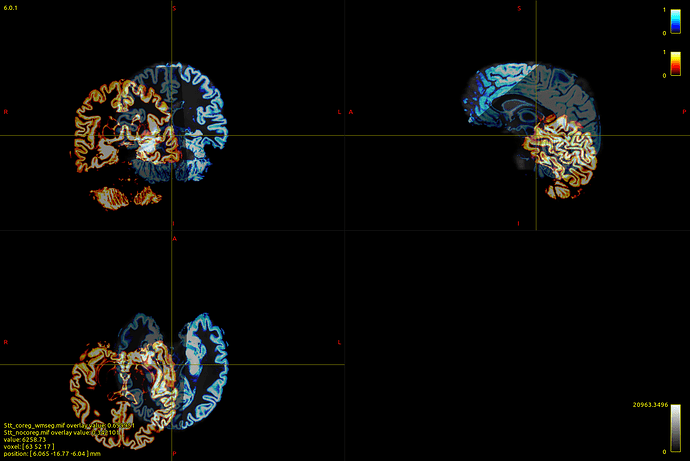Dear MRtrix3 users and developers,
I’m trying to adapt the preprocessing pipeline available at OSF (B.A.T.M.A.N) to my data in order to perform Anatomically Constrained Tractography (OSF | B.A.T.M.A.N.: Basic and Advanced Tractography with MRtrix for All Neurophiles). Unfortunately I encounter some form of image registration error between T1 and DWI images.
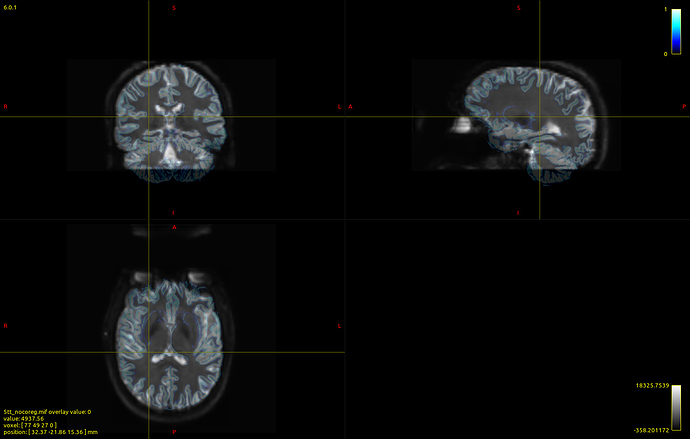
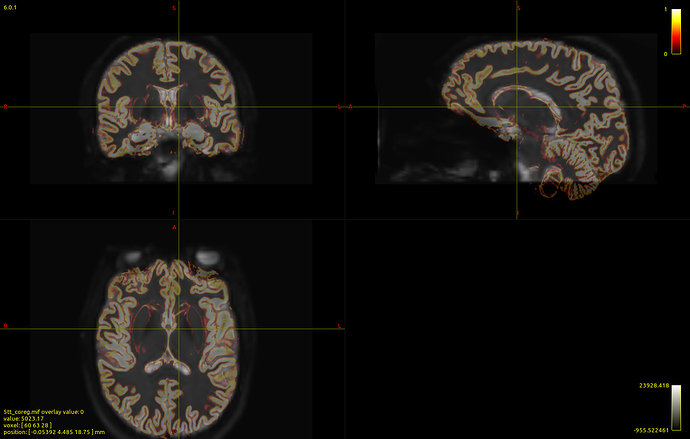
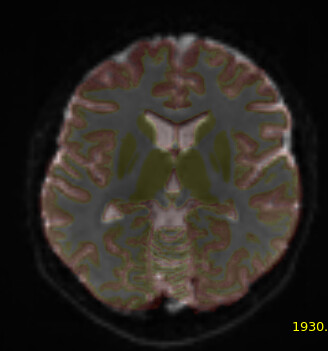
Here’s a picture of my dwi_b0 image with the 5tt_nocoreg.mif image overlayed onto it (almost perfectly aligned):
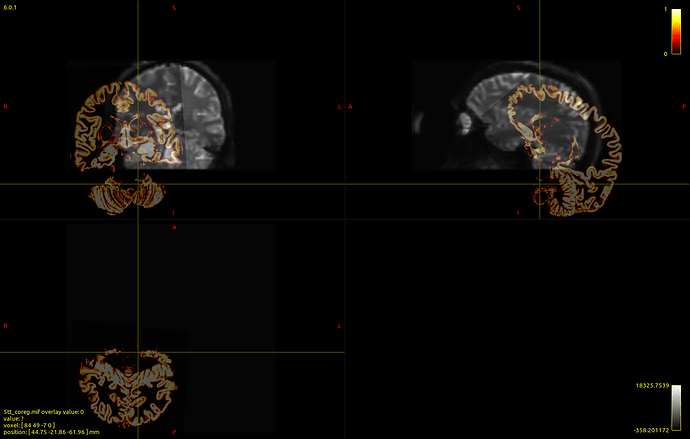
Here’s a picture of my dwi_b0 image with the 5tt_nocoreg.mif image overlayed onto it (crazy stuff here!):
Here’s my bit of code for five tyssue type generation and then registration to dwi images:
…
#converting DICOM T1 directory to T1 image in .mif format
mrconvert /T1 T1_raw.mif
#preparing data for anatomically constrained tractography (ACT) to increase biological plausibility of stramlines
5ttgen fsl T1_raw.mif 5tt_nocoreg.mif -nocleanup
#compute mean of b0 images
dwiextract dwi_den_unr_preproc.mif - -bzero | mrmath - mean mean_b0_preprocessed.mif -axis 3
mrconvert mean_b0_preprocessed.mif mean_b0_preprocessed.nii.gz
bet mean_b0_preprocessed.nii.gz mean_b0_preprocessed_BET.nii.gz
#getting images for registration
cp 5ttgen-tmp-*/T1_BET.nii.gz ./
cp 5ttgen-tmp-*/T1_BET_pve_2.nii.gz ./
mrconvert T1_raw.mif T1.nii.gz
#register dwi to T1 and then apply inverse transform to register 5tt image to dwi
epi_reg --epi=mean_b0_preprocessed_BET.nii.gz --t1=T1.nii.gz --t1brain=T1_BET.nii.gz --wmseg=T1_BET_pve_2.nii.gz --out=diff2struct_fsl
transformconvert diff2struct_fsl.mat mean_b0_preprocessed_BET.nii.gz T1.nii.gz flirt_import diff2struct_mrtrix.txt
mrtransform 5tt_nocoreg.mif -linear diff2struct_mrtrix.txt -inverse 5tt_coreg.mif
#check results with:
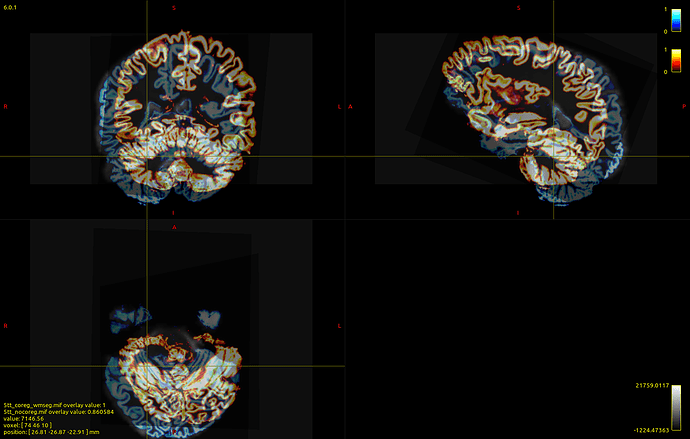
mrview dwi_den_unr_preproc.mif -overlay.load 5tt_nocoreg.mif -overlay.colourmap 2 -overlay.load 5tt_coreg.mif -overlay.colourmap 1
Here there is mrinfo’s output of 5tt_nocoreg.mif and 5tt_coreg.mif
> ************************************************
> Image: "5tt_nocoreg.mif"
> ************************************************
> Dimensions: 192 x 245 x 195 x 5
> Voxel size: 0.700521 x 0.700521 x 0.7 x ?
> Data strides: [ 2 3 4 1 ]
> Format: MRtrix
> Data type: 32 bit float (little endian)
> Intensity scaling: offset = 0, multiplier = 1
> Transform: 0.9998 -0.01169 0.01318 -59.43
> 0.01551 0.9388 -0.3442 -70.12
> -0.008352 0.3444 0.9388 -79.66
> command_history: /home/duzzo/mrtrix3/bin/mrcalc "T1_BET_pve_1.nii.gz" "multiplier_noNAN.mif" "-mult" "remove_unconnected_wm_mask.mif" "-mult" "cgm.mif" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
> /home/duzzo/mrtrix3/bin/mrcat "cgm.mif" "sgm.mif" "wm.mif" "csf.mif" "path.mif" "-" "-axis" "3" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
> /home/duzzo/mrtrix3/bin/mrconvert "-" "combined_precrop.mif" "-strides" "+2,+3,+4,+1" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
> /home/duzzo/mrtrix3/bin/mrcrop "combined_precrop.mif" "result.mif" "-mask" "-" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
> /home/duzzo/mrtrix3/bin/mrconvert "result.mif" "/mnt/226AE1226AE0F407/DTI/pipeline_test/Duzzo/5tt_nocoreg.mif" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
> comments: 6.0.1
> mrtrix_version: 3
************************************************
Image: "5tt_coreg.mif"
************************************************
Dimensions: 192 x 195 x 245 x 5
Voxel size: 0.700521 x 0.7 x 0.700521 x ?
Data strides: [ 2 -4 3 1 ]
Format: MRtrix
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 0.9962 -0.08495 0.01874 -3.205
0.05217 0.7558 0.6527 -155.2
-0.06962 -0.6493 0.7574 -35.24
command_history: /home/duzzo/mrtrix3/bin/mrcalc "T1_BET_pve_1.nii.gz" "multiplier_noNAN.mif" "-mult" "remove_unconnected_wm_mask.mif" "-mult" "cgm.mif" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
[6 entries] /home/duzzo/mrtrix3/bin/mrcat "cgm.mif" "sgm.mif" "wm.mif" "csf.mif" "path.mif" "-" "-axis" "3" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
...
/home/duzzo/mrtrix3/bin/mrconvert "result.mif" "/mnt/226AE1226AE0F407/DTI/pipeline_test/Duzzo/5tt_nocoreg.mif" "-nthreads" "4" (version=3.0_RC3-137-g5d6b3a6f)
mrtransform "5tt_nocoreg.mif" "-linear" "diff2struct_mrtrix.txt" "-inverse" "5tt_coreg.mif" (version=3.0_RC3-137-g5d6b3a6f)
comments: 6.0.1
transform modified
mrtrix_version: 3.0_RC3-137-g5d6b3a6f
.0_RC3-137-g5d6b3a6f
Does anyone have an idea of why this is happening or where my pipeline is wrong?
thanks for your kind help and support
Davide