Hi @nayan_wadhwani,
Apologies, I’m not familiar enough with the specific conventions assumed when storing streamlines in VTK format to be able to give you a definite answer, but I can probably give you some pointers as to what might be going on.
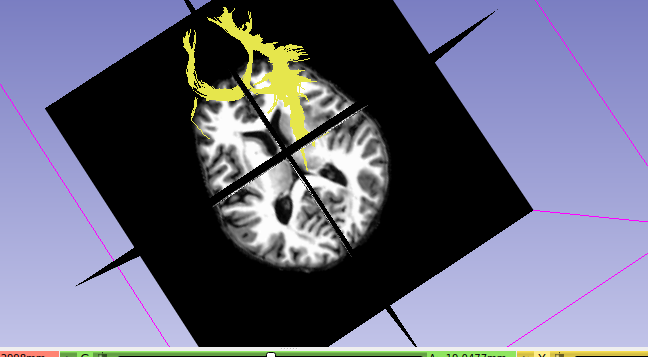
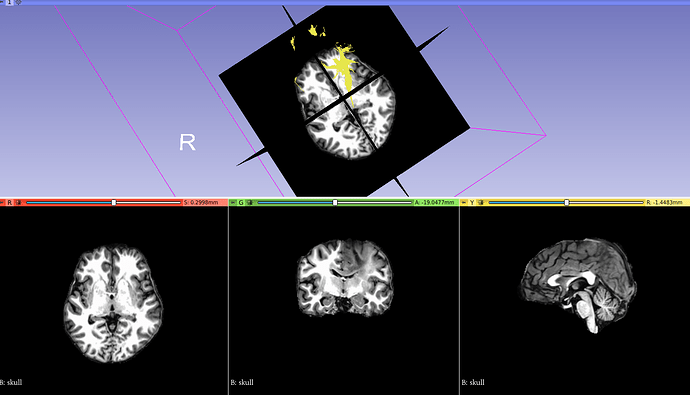
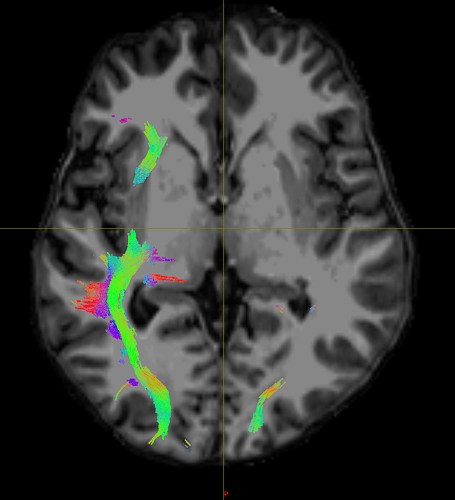
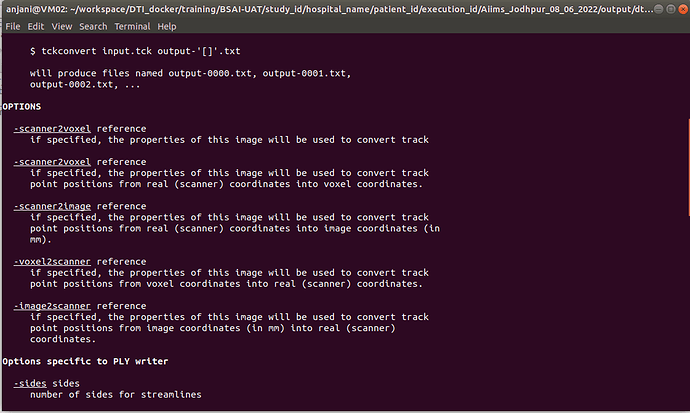
In TCK, the position of each vertex (a 3D point along the streamline) is stored relative to the real / world / scanner coordinate system (essentially the XYZ axes of the scanner). In other formats, there’s a good chance the positions are stored in voxel coordinates relative to the axes of the diffusion MRI dataset they were generated from – I expect this might be the case with the VTK format (though I’m struggling to find concrete documentation on this). This means that to figure out the coordinates of each vertex in the VTK file, you need to provide a reference image whose metadata provides the information required to work out the mapping between scanner and voxel coordinates (typically the voxel sizes, image transform, image dimensions). This information would need to be provided to the tckconvert call, most likely via the -scanner2voxel option¹.
This might be enough to fix the issue, but you then have the corresponding problem when loading the data into other packages like Slicer. I’m not familiar with this package, so I’m not sure how it handles data of this nature. But to be able to properly handle the VTK data, it would also need to know which coordinate system to assume when reading the data. I don’t think the VTK file contains any information about this (at least, the VTK files produced by tckconvert don’t), so the reference image would also need to be provided to Slicer for it to be able to work out where to put each vertex in real / scanner / world space (assuming that’s the coordinate system it uses internally, which might not be the case).
The situation is likely to get more complicated again if you’re trying to overlay the streamlines generated from a diffusion MRI dataset (with its own orientation, voxel size, and dimensions) on top of a different image, such as the anatomical (which will have its own different orientation, voxel size and dimensions). Again, it depends on how Slicer operates, but it could be that it then needs to figure out the transformation from the diffusion image to world coordinates, and then from world coordinates to the anatomical image. I have no idea how it handles this situation… Maybe it’s sufficient to provide the anatomical image to tckconvert -scanner2voxel, and then everything works out once in Slicer, but that would need testing.
Note that we avoid these issues in MRtrix by explicitly storing everything in world coordinates – no need for an external reference image, it’s completely unambiguous. This was an early design decision that I think has proved its worth…
Hope this helps,
Donald
¹though possibly using the -scanner2image option if vertices are stored in the image frame, but in millimeter units rather than voxel units – again, I can’t find solid documentation on this…