Hi all,
I am trying to generate structural connectome between bilateral motor areas using ROIs defined by fMRI.
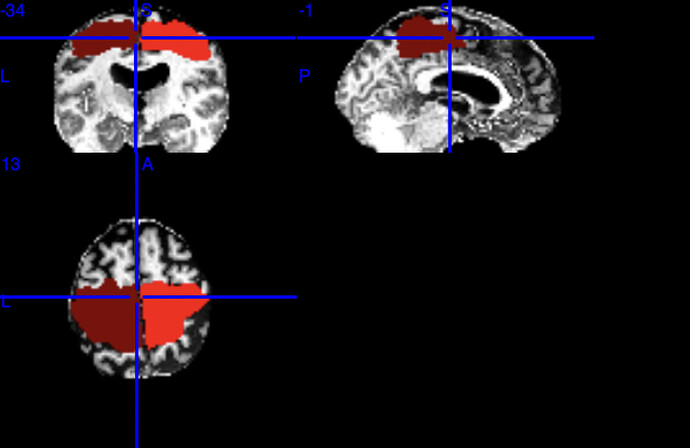
The ROIs are as following, where left is labelled 1 and right labeled 2:
Then I tried to extract connections between these two regions using the following command:
tck2connectome -symmetric -zero_diagonal -tck_weights_in sift_1M.txt tracks_10M.tck Moto.mif Moto.csv -out_assignment assignment_Moto.csv
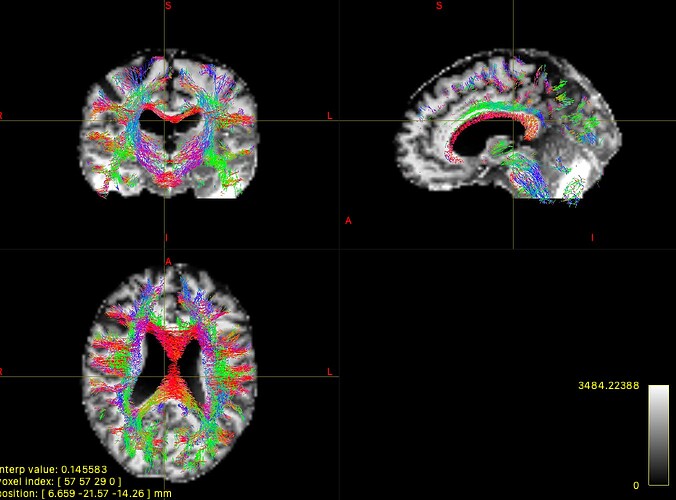
However, when i tried to visualize the generated tracks it seems there are way more tracks than I expected:
connectome2tck –nodes 1,2 –exclusive tracks_10M.tck -tck_weights_in sift_1M.txt assignments_Moto.csv moto
While I only expected the bilateral motor areas were connected by the corpus callosum, it seems that in the image there are tracks basically anywhere in the brain. Did I do something wrong?
Thank you very much!
Best
Xinyu
Hi Xinyu,
It might be worth checking the tck2connectome inputs, in particular, the -tck_weights_in and the corresponding .tck file. It looks like the weights file belongs to a .tck file containing 1M streamlines, and the input .tck file contains 10M streamlines… was there a warning or error message when running this command?
Cheers,
Arkiev
Hi Arkiev,
No there is no error message. I did this because
tck2connetome
requires a .tck file as input, but
tcksift2
generates a .txt file. Could you guide me how to do it correctly? I tried only using .tck with 10M streamlines as input but the result is similar.
Dear Xinyu,
As far as I understood, you are interested in bilateral motor cortices connections in your study. I wonder why you haven’t done direct tractography between these two regions, and have decided to go with tck2connectome and after that connectome2tck. In general, the guideline is to do both global and ROI-ROI tractography, and combine them together.
Anyway, getting back to your question:
- Your motor cortices are too large, increasing the chances of including so many tracts
- You have to set a minimum length or whatever for your tracks, or exclude masks
- You have to check whether 1 and 2 really assign to the motor cortices, it is possible that nodes 1 and 2 are maybe the whole white matter regions or whatever
- Follow the “EXAMPLE USAGES” of connectome2tck; I expect the command you’ve written will generate errors about that “assignments_Moto.csv” thing\
Best,
Amir