Dr. Tournier, thank you very much for replying and offering to help.
To clarify the issue I am having, I have run tckgen on the same masks and FOD, using the same command, on two different machines. Machine A is my local machine, using MRtrix 3, on the May 4 2018 release. Machine B is the HPC cluster, running a more updated version of MRtrix 3 released on March 3, 2019. I am trying to identify tracts between the ZI and PVT in HCP subjects. I ran one subject on Machine A, and then wanted to run on Machine B. Even after copy and pasting the FOD and masks from Machine A to Machine B, and running the same command, I got drastically different results.
This is the command I used:
tckgen -info FOD_WM.mif lh_zi_to_pvt_2.tck -seed_image sub-63_lh_zi_dwi.nii.gz -force -include sub-63_lh_pvt_dwi.nii.gz -include sub-63_lh_zi_dwi.nii.gz -seeds 1065000 -select 0
These are the outputs of mrinfo:
MacBook-Pro-172:sub-63 sabir$ mrinfo FOD_WM.mif
************************************************
Image: "FOD_WM.mif"
************************************************
Dimensions: 173 x 207 x 173 x 45
Voxel size: 1.05 x 1.05 x 1.05 x 1
Data strides: [ -2 3 4 1 ]
Format: MRtrix
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 -90.6
-0 1 0 -126
-0 0 1 -72
command_history: mrconvert "sub-63_dwi.nii.gz" "-fslgrad" "sub-63_dwi.bvec" "sub-63_dwi.bval" "sub-63_dwi.mif" (version=e27b1200)
dwi2fod "msmt_csd" "sub-63_dwi.mif" "response_wm.txt" "FOD_WM.mif" "response_gm.txt" "FOD_GM.mif" "response_csf.txt" "FOD_CSF.mif" "-mask" "sub-63_dwi_bet_mask.nii.gz" (version=e27b1200)
comments: FSL5.0
mrtrix_version: e27b1200
prior_dw_scheme: -0.02906800072,0.9090760224,-0.4156150103,0
[143 entries] -0.5324862053,0.05988902309,0.8443173255,979.9992444
...
-0.6565713652,-0.4873472711,0.5756793202,984.9989042
-0.4038150334,0.4291610355,0.8079320667,2019.999666
MacBook-Pro-172:sub-63 sabir$ mrinfo sub-63_lh_zi_dwi.nii.gz
************************************************
Image: "sub-63_lh_zi_dwi.nii.gz"
************************************************
Dimensions: 173 x 207 x 173
Voxel size: 1.05 x 1.05 x 1.05
Data strides: [ -1 2 3 ]
Format: NIfTI-1.1 (GZip compressed)
Data type: 64 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 -90.6
-0 1 0 -126
-0 0 1 -72
comments: 5.0.10
MacBook-Pro-172:sub-63 sabir$ mrinfo sub-63_lh_pvt_dwi.nii.gz
************************************************
Image: "sub-63_lh_pvt_dwi.nii.gz"
************************************************
Dimensions: 173 x 207 x 173
Voxel size: 1.05 x 1.05 x 1.05
Data strides: [ -1 2 3 ]
Format: NIfTI-1.1 (GZip compressed)
Data type: 64 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 -90.6
-0 1 0 -126
-0 0 1 -72
comments: 5.0.10
The reason I suspected the issue to arise in tckgen is because the FOD (and everything else) was the same. I used the -info flag in tckgen to see what was happening in each step. In Machine A, there was no message that said there was reorientation of the axes, but in Machine B there was.
Machine A -info:
tckgen: [WARNING] existing output files will be overwritten
tckgen: [INFO] opening image "sub-63_lh_pvt_dwi.nii.gz"...
tckgen: [INFO] image "sub-63_lh_pvt_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: [100%] uncompressing image "sub-63_lh_pvt_dwi.nii.gz"
tckgen: [INFO] opening image "sub-63_lh_zi_dwi.nii.gz"...
tckgen: [INFO] image "sub-63_lh_zi_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: [100%] uncompressing image "sub-63_lh_zi_dwi.nii.gz"
tckgen: [INFO] opening image "sub-63_lh_pvt_dwi.nii.gz"...
tckgen: [INFO] image "sub-63_lh_pvt_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: [100%] uncompressing image "sub-63_lh_pvt_dwi.nii.gz"
tckgen: [INFO] opening image "sub-63_lh_zi_dwi.nii.gz"...
tckgen: [INFO] image "sub-63_lh_zi_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: [100%] uncompressing image "sub-63_lh_zi_dwi.nii.gz"
tckgen: [INFO] opening image "sub-63_lh_zi_dwi.nii.gz"...
tckgen: [INFO] image "sub-63_lh_zi_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: [100%] uncompressing image "sub-63_lh_zi_dwi.nii.gz"
tckgen: [INFO] opening image "FOD_WM.mif"...
tckgen: [INFO] image "FOD_WM.mif" opened with dimensions 173x207x173x45, voxel spacing 1.05x1.05x1.05x1, datatype Float32LE
tckgen: [INFO] step size = 0.524999976 mm
tckgen: [INFO] maximum deviation angle = 45 deg
tckgen: [INFO] minimum radius of curvature = 1.0499999230973744 mm
tckgen: [INFO] iFOD2 internal step size = 0.174999997 mm
tckgen: [INFO] rejection sampling will use 7 directions with a ratio of 2.15373945 (predicted number of samples per step = 12.9595909)
Machine B -info:
tckgen: [WARNING] existing output files will be overwritten
tckgen: [INFO] opening image "sub-63_lh_pvt_dwi.nii.gz"...
tckgen: [INFO] Axes and transform of image "sub-63_lh_pvt_dwi.nii.gz" altered to approximate RAS coordinate system
tckgen: [INFO] image "sub-63_lh_pvt_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: uncompressing image "sub-63_lh_pvt_dwi.nii.gz"... [================================]
tckgen: [INFO] opening image "sub-63_lh_zi_dwi.nii.gz"...
tckgen: [INFO] Axes and transform of image "sub-63_lh_zi_dwi.nii.gz" altered to approximate RAS coordinate system
tckgen: [INFO] image "sub-63_lh_zi_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: uncompressing image "sub-63_lh_zi_dwi.nii.gz"... [================================]
tckgen: [INFO] opening image "sub-63_lh_pvt_dwi.nii.gz"...
tckgen: [INFO] Axes and transform of image "sub-63_lh_pvt_dwi.nii.gz" altered to approximate RAS coordinate system
tckgen: [INFO] image "sub-63_lh_pvt_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: uncompressing image "sub-63_lh_pvt_dwi.nii.gz"... [================================]
tckgen: [INFO] opening image "sub-63_lh_zi_dwi.nii.gz"...
tckgen: [INFO] Axes and transform of image "sub-63_lh_zi_dwi.nii.gz" altered to approximate RAS coordinate system
tckgen: [INFO] image "sub-63_lh_zi_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: uncompressing image "sub-63_lh_zi_dwi.nii.gz"... [================================]
tckgen: [INFO] opening image "sub-63_lh_zi_dwi.nii.gz"...
tckgen: [INFO] Axes and transform of image "sub-63_lh_zi_dwi.nii.gz" altered to approximate RAS coordinate system
tckgen: [INFO] image "sub-63_lh_zi_dwi.nii.gz" opened with dimensions 173x207x173, voxel spacing 1.0499999523162842x1.0499999523162842x1.0499999523162842, datatype Float64LE
tckgen: uncompressing image "sub-63_lh_zi_dwi.nii.gz"... [================================]
tckgen: [INFO] opening image "FOD_WM.mif"...
tckgen: [INFO] image "FOD_WM.mif" opened with dimensions 173x207x173x45, voxel spacing 1.05x1.05x1.05x1, datatype Float32LE
tckgen: [INFO] step size = 0.524999976 mm
tckgen: [INFO] maximum deviation angle = 45 deg
tckgen: [INFO] minimum radius of curvature = 1.0499999230973744 mm
tckgen: [INFO] iFOD2 internal step size = 0.174999997 mm
tckgen: [INFO] rejection sampling will use 7 directions with a ratio of 2.15373945 (predicted number of samples per step = 12.9595909)
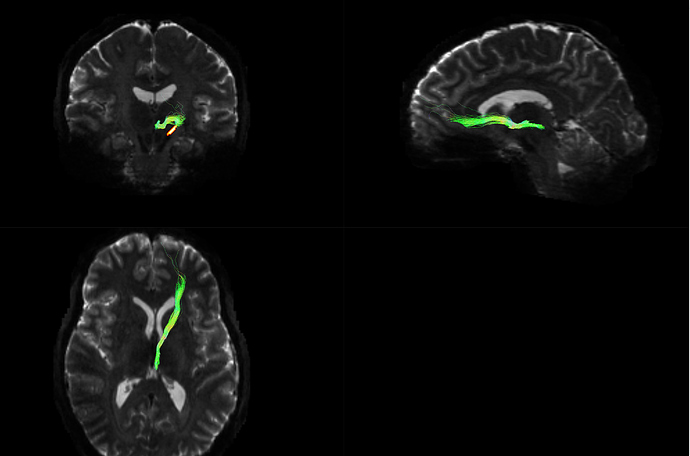
Machine A output tck:
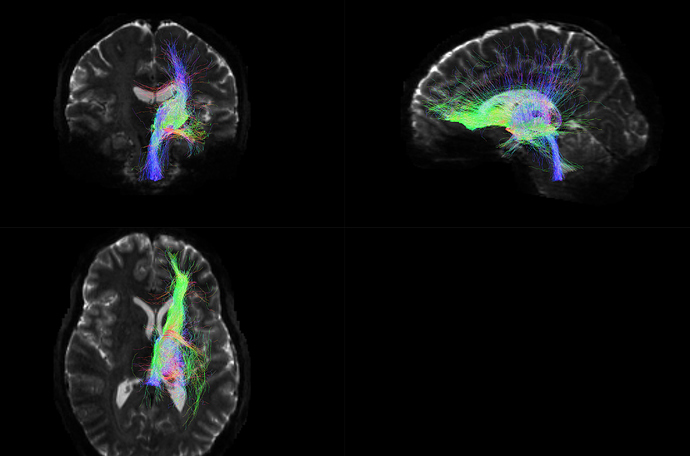
Machine B output tck:
(Visualizing both of these images in MRview, unchecked the “crop to slab” option)
These are the commands i used to generate the FOD.
bet sub-63_dwi.nii.gz sub-63_dwi_bet.nii.gz
mrconvert sub-63_dwi.nii.gz -fslgrad sub-63_dwi.bvec sub-63_dwi.bval sub-63_dwi.mif
dwi2response dhollander sub-63_dwi.mif response_wm.txt response_gm.txt response_csf.txt -mask sub-63_dwi_bet.nii.gz
dwi2fod msmt_csd sub-63_dwi.mif response_wm.txt FOD_WM.mif response_gm.txt FOD_GM.mif response_csf.txt FOD_CSF.mif -mask sub-63_dwi_bet_mask.nii.gz
So, yeah. I’m kind of stuck. If you have any insight on how to resolve this, I would really appreciate it. Thanks again for the help.
-Sabir