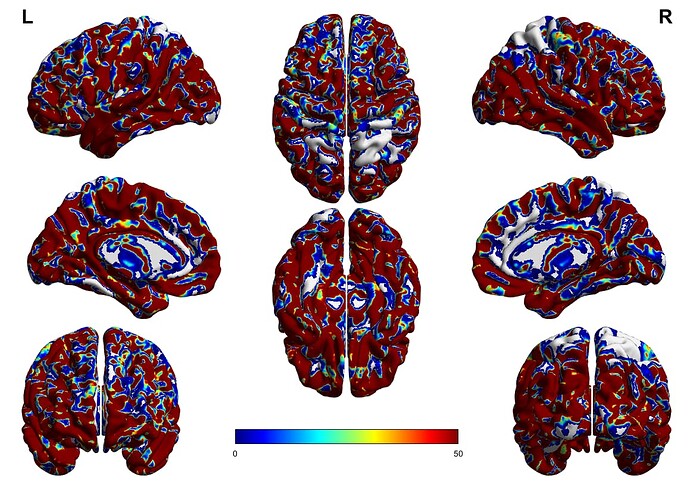
Hey All,
So I have been conducting whole brain tractography, some subjects I get a pretty good coverage of the brain but in other subjects (such as the one shown below) when I look at the tckmap (mapping the endpoints of the tractography) there are some areas that in fact do not have tractography endpoints (noted by grey/white coloration). Why would this be the case? The first came to mind was to up the number of streamlines selected in the tckgen algorithm-which remedied it somewhat, but still resulted in these regions without tracts ending. Could this potentially be a result of an underlying registration issue?
I should note that this is a healthy control too!
Cheers,
Ricardo
Hi Ricardo,
It’s exceptionally difficult to draw any conclusions about the results in isolation, especially for the cerebellar data, as it’s unclear how the projection is being generated. I’d need to know every detail about the reconstruction, and probably be looking at the raw data of every step very closely.
Homogeneity of coverage at the cortex is highly dependent on the seeding strategy. Homogeneous WM seeding in particular will produce hotspots, and probably cold spots; not sure what would be more homogeneous between interface and dynamic seeding. SIFT(2) will also decrease erroneous variance to some extent. This manuscript is worth reading on the topic.
You could perform targeted tracking, seeding in a region not covered by the whole-brain tractogram, and see where any generated streamlines go / why those streamlines are deemed unacceptable (using tckgen -info). Sometimes a gyrus can look almost “cut off” from the rest of the parenchyma due to very narrow superficial WM, which tractography will struggle to pass through reliably, and streamlines could then be rejected if the minimum length is too long.
There could also be an issue with the way in which the streamlines are projected up to the surface. E.g. You could have a slight mismatch between where the streamlines are terminating during propagation vs. the surface to which they are assigned, resulting in a cold spot just due to the processing pipeline not being adequately robust.
So I can only really suggest looking at all of the intermediate data very closely, rather than trying to hypothesize based only on the end result.
Cheers
Rob
hello, thank you for your insight. Going back and visualizing the results through mrview we concluded it was the surface files from the third party viewer (brainnetviewer) All of the processing was confined to and projected onto the gmwmi. All of the surfaces available via brainnet are mostly representative of cortical layers and can be quiet tricky to showcase the gmwmi. As a results the results are being displayed via lightbox in mrview and they look so much better. thanks for the help!