Hi all,
I have a question regarding the “FA weighted connectome”.
I do the following:
tcksample $OUTDIR/Tractogram.tck $OUTDIR/FA.nii.gz $OUTDIR/FA.csv -stat_tck mean -force
tck2connectome $OUTDIR/Tractogram.tck $OUTDIR/filtered_GM_parcel_adapted.nii.gz $OUTDIR/Connectivity_matrix_FA.csv -scale_file $OUTDIR/FA.csv -stat_edge mean -force
But now I was wondering what happens if for example the following situation happens:
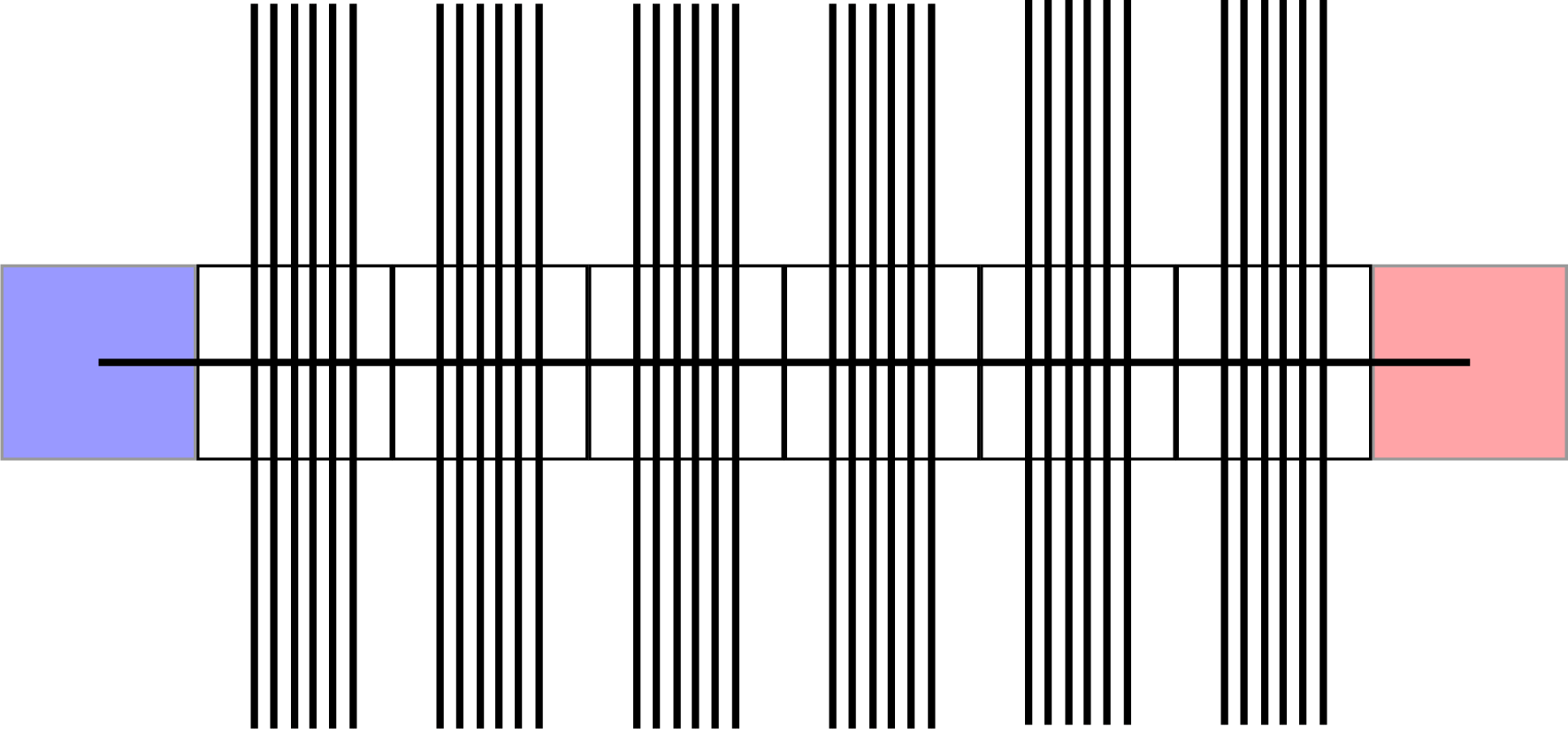
So region Blue is connected with region Red via 1 streamline that crosses multiple voxels with a lot of streamlines vertically. Because of the vertically oriented streamlines these voxels have for example an FA of 0.6. Now will this single streamline have an average FA of 0.6 over the whole length? Because the 0.6 in these voxels is completely caused by fibers oriented in a different direction.
I think concise my main question is:
Does it take into account the directionality or for examples FODs in the voxels or is there a way that this is possible?
Thanks in advance! Boaz