Thanks for taking the time to do this, it’s appreciated.
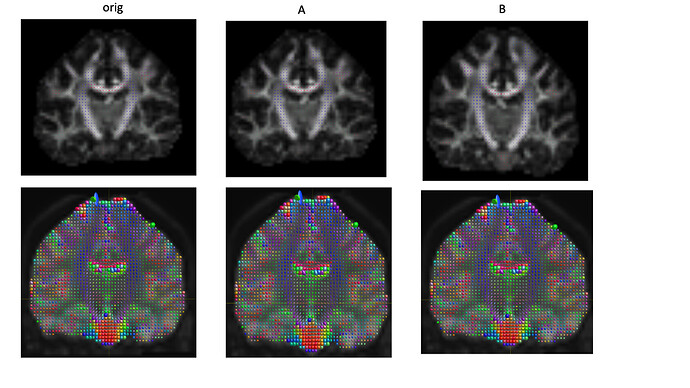
OK, assuming the top row is TORTOISE, and the bottom row is MRtrix, the issue matches what I expected. It’s far from trivial unfortunately, due to the idiosyncrasies of the bvecs/bvals format. The main issue relates to this small, but important addition to the FSL wiki where they describe how to interpret these files (my emphasis):
The bvecs use a radiological voxel convention, which is the voxel convention that FSL uses internally and originated before NIFTI (i.e., it is the old Analyze convention). If the image has a radiological storage orientation (negative determinant of qform/sform) then the NIFTI voxels and the radiological voxels are the same. If the image has a neurological storage orientation (positive determinant of qform/sform) then the NIFTI voxels need to be flipped in the x-axis (not the y-axis or z-axis) to obtain radiological voxels.
An image stored with strides 1,2,3,4 is stored using the standard NIfTI coordinate system, which is ‘neurological’ (positive qfac / positive determinant of the qform/sform) – whereas an image with strides -1,2,3,4 is ‘radiological’ (note that I really dislike the use of these terms in this context, I would normally use neurological & radiological to refer to the display of the images only, not how they’re stored on disk). This means that the x component of the bvecs needs to be flipped (inverted) if the image it relates to is stored this way.
Unfortunately, as far as I can tell, this means that the data passed to TORTOISE is not actually compliant with FSL – it would be good if you could confirm that by running FSL’s dtifit and looking at the results in fslview or fsleyes. I’m pretty confident our bvecs handling is consistent with FSL (which is the reference implementation for that format), so I think the issue here is that TORTOISE is not handling these files as required. It would be good to confirm this against FSL and report the issue to the TORTOISE developers.
Given that these data were converted using dcm2niix, I’d also recommend getting @Chris_Rorden’s opinion as to what might be going on here. As I understand it, dcm2niix doesn’t try to reorder the data, so I would expect the images produced to have strides -1,-2,3,4 – corresponding to their ordering in the original DICOM data. I suspect some other operation has been applied to the data to reorient it to the standard NIfTI frame, was this the case? In which case, there’s the possibility that this operation might have introduced some discrepancy here?
 …
…