Hi everyone,
I have a question regarding masks and image thresholding. I used freesurfer to create ROI masks and co-registered the masks with mrtransform to my FA maps with nearest-neighbor interpolation. My problem is that the interpolation seems to increase the mask size.
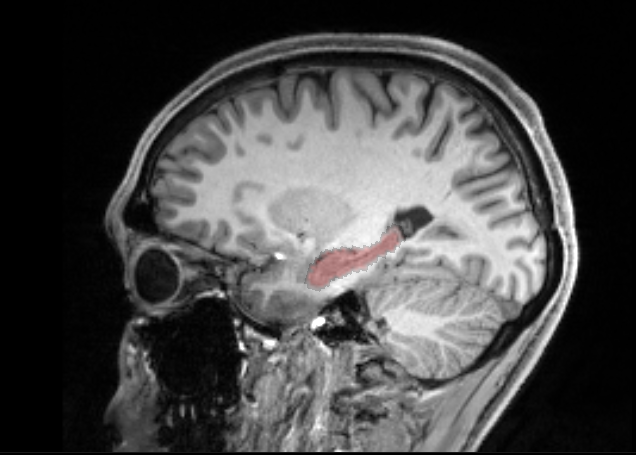
By coincidence, I realized in mrview that by overlaying a T1 with the ROI mask, I can apply a threshold in the viewer (see image) that drastically improves the mask.
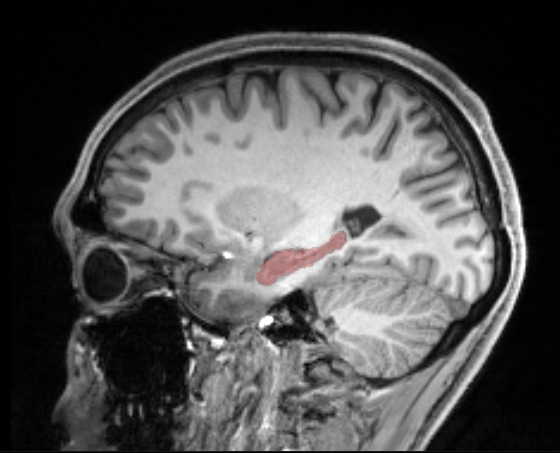
However, there seems to be no way of exporting the mask with the threshold.
I already tried using mrtrix commands for this, in particular mrthreshold, but I this does not work at all. So my question is: is there any mrtrix command that corresponds to the overlay threshold in mrview?
Thank you for your help!
Best
Lars
Hi Lars,
I assume using trilinear interpolation instead of nearest neighbor and thresholding the result with 0.5 using mrthreshold would give you the desired result
BW
William
1 Like
Hi William,
thank you very much for your reply! In fact, I realized that the thresholding method won’t work in any kind of way, because my brain mask is binarized (I just forgot about it). It seems that mrview did some kind of interpolation when I overlayed the T1 with the ROI mask, which non-binarized the mask at its border.
So far, the best solution was to either (1) apply mrfilter with gaussian smoothing (which I guess it similar to what mrview is doing) and then apply mrthreshold or (2) simply use MRtrix’ maskfilter with the erode function and npass = 1. Both worked fine, but I decided for the latter for its simplicity.
Anyway, thanks for your help!
Best
Lars
Hi Lars,
Solution one is similar to what I proposed i.e., using trilinear interpolation when transforming your ROI from freesurfer to your FA maps and then thresholding. Your second solution “eats some” voxels away. It depends on what you want to do with the ROIs and the type of ROI, I guess. Solution 2 may decrease the possible contribution of WM and CSF voxels. However, if minimizing the contribution of none GM to the hippocampus is your goal, you might want to do some additional “cleaning” using FA and MD thresholding
BW
William
1 Like
Thank you very much for your explanation! In fact, decreasing WM / CSF influence is indeed one of the goals (in addition of course to sticking to the ideal of “pure” hippocampal voxels). I will definitely consider your suggestion regarding additional cleaning with an FA threshold.
Best
Lars
Hi Lars,
I think the easiest way is to uncheck the interpolate box as shown below.
Have a try!
Best,
Volcano
Hi William,
I have tried your solution with trilinear interpolation, because it seems ideal for my case. The problem I have is, that FSL is doing weird stuff with my brain masks. Currently, these are my steps:
- Convert Freesurfer output aparc+aseg.mgz to aparc+asec.nii
- Extract hippocampus with: mri_binarize --i aparc+aseg.nii --o mask --match 17
- Co-register and interpolate hippocampus mask to diffusion space with inverted transformation matrix from FSL epireg: flirt -in l_hippocampus_mask.nii.gz -ref fa_map.nii.gz -applyxfm -interp trilinear -init struct2diff.mat -out l_hippocampus_mask_coreg_interpol.nii.gz
Unfortunately, flirt is doing a terrible job here, and moves my mask to a random location within the brain. If I use the same transformation matrix for the entire aparc+aseg, it works just fine, but then I can’t extract the hippocampus anymore due to the interpolation.
The weird thing is, that all these steps work perfectly fine in MRtrix, but MRtrix does not seem to have the trilinear interpolation algorithm, but only cubic interpolation (or is this just a different name for the same thing?).
Do you have any suggestions what might go wrong here?
Best
Lars
Hi Volcano,
In my case, I am not having issues with the viewer, but with optimizing my brain masks for later analysis. Still, thank you very much for taking the time to reply!
Best
Lars
Hi Lars,
Assuming, you use mrtransform in MRtrix, it has the option:
-interp method set the interpolation method to use when reslicing (choices: nearest, linear, cubic, sinc. Default: cubic)
you should then use “linear”
Hope this helps
Cheers
William
1 Like
Ah, I thought linear would be different from trilinear. Thank you so much for your help!
Best
Lars