Dear MRtrix team,
I have a question regarding the method described in your paper to perform tractography in patients with intrinsic brain tumours:
I have patients with tumours for which I performed the following steps.
-
5tt fsl segmentation on an enantiomorphically filled T1 with tumour masking as pathological tissue in the final 5TT image.
-
single shell 3-tissue CSD to generate RFs.
-
mtnormalise
-
tckgen -act $t1 -backtrack -seed_dynamic $wm -select 50M -maxlength 250 -cutoff 0.06 $wm 50M.tck -info
-
tcksift2 -act $t1 50M.tck $wm 50M.sift -out_mu SIFT2_mu.txt -out_coeffs tck_coeffs.txt -info
-
generation of TDI.
In the paper’s description of the method, it states that:
'resulting white matter FOD amplitudes were substantially smaller in tumor regions compared to the rest of the brain. While this likely reflects the smaller portion of space taken up by axons (due to infiltrating tumor tissue) and/or damage to white matter tracts, it does pose a practical challenge to tractography algorithms, which rely on the aforementioned amplitude threshold to determine where and how far tractography may proceed. We overcame this by gradually reducing the FOD amplitude threshold close to and even more so within the tumor, based on its prior segmentation obtained registering the T1-weighted image to the diffusion data’
I am unsure how this modulation is achieved?
I cannot see any options in tckgen which allows you to change the FOD threshold based on a tumour mask for example? In addition, although these tumours can be structurally classed as pathological tissue to avoid mislabelling as GM and streamline termination. Large portions of these tumours have RFs which resemble CSF and GM (likely expected due to their heterogenous and cystic natures).
I have included an example 10M streamline tractography output and corresponding TDI below:
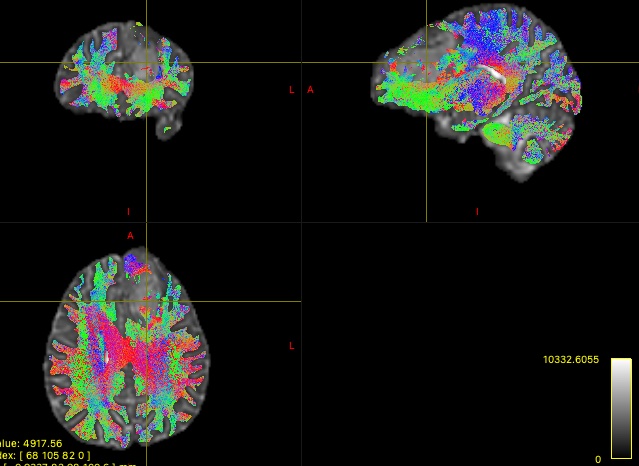
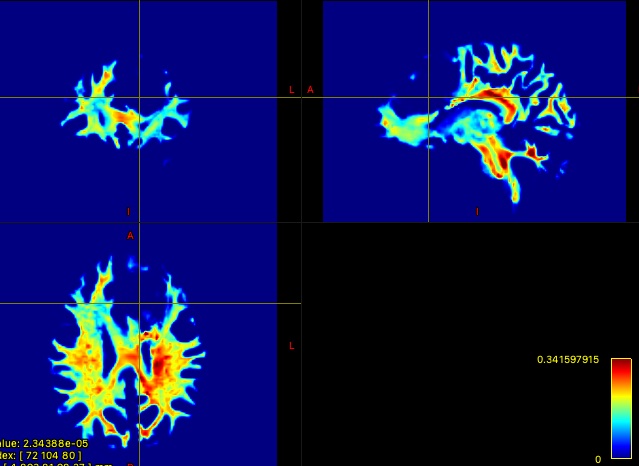
Thanks!
Yizhou
1 Like
Hi Yizhou,
Pinging @Hannelore_Aerts and @bjeurissen since I don’t always have all the answers 
You’re correct in that tckgen does not currently have any mechanism by which to spatially modulate the FOD amplitude threshold. It is however something that is obviously technically feasible, albeit potentially clumsy from a command interface perspective. If this is a feature that you would make use of were it implemented in MRtrix3, I would suggest posting it as a feature request on GitHub. We can then see if the modifications that were done for the purpose of that manuscript can be integrated into the distributed code, or anybody interested in investing effort in implementing that functionality can get technical advice from the developers.
Also, just quickly:
- single shell 3-tissue CSD to generate RFs.
This conflation seems to occur very regularly, and I’m not quite sure how best to address it. Single-shell 3-tissue CSD (and equivalently other CSD variants) is used to generate ODFs, which are calculated based on the RFs. The RFs are generated using a response function estimation algorithm; which, though some of those algorithms may make use of CSD within their internal optimisation processes, are not themselves “CSD”, but are better thought of as precursors to CSD.
- generation of TDI.
From your provided images (based on the inhomogeneity throughout the WM) I’m guessing that you did not provide the SIFT2 weights from step 5 when the TDI was generated in step 6. That’s totally legitimate, it just renders step 5 redundant in the explanation of your steps, which is slightly misleading.
In addition, although these tumours can be structurally classed as pathological tissue to avoid mislabelling as GM and streamline termination. Large portions of these tumours have RFs which resemble CSF and GM (likely expected due to their heterogenous and cystic natures).
The volumes in the 5TT image can be derived from any information you choose. Further, they don’t necessarily have to be treated as “this is what this tissue is”; it can rather be “these are the anatomical priors that I want ACT to apply in this location”. All the “pathological tissue” class in the 5TT image does is disable the anatomical priors in that location (and indeed I probably chose the wrong noun for that 5th volume).
So for instance, you could say something like: “In this location, I don’t trust the outcomes of tissue segmentation based on the T1-weighted image; but I do think that the outcomes of DWI-based tissue decomposition are reasonable”; at which point you could integrate the signal fractions from multi-tissue CSD into your 5TT image and use those to drive the ACT anatomical priors just within the pathological region. I thought about this years ago, but have never had an application on my desk to justify the investment of time writing a script to do it.
(Admittedly such an approach might not behave exactly as desired since ACT is designed around hard tissue borders rather than diffuse mixtures of tissues, but there’s certainly room for experimentation)
Cheers
Rob
Hi Yizhou,
I don’t use this forum frequently anymore these days; somebody highlighted your question directly to me just now. As the work indeed states and cites:
As detailed in Aerts et al. (2019), we overcame this by gradually reducing the FOD amplitude threshold close to and even more so within the tumor, based on its prior segmentation obtained registering the T1-weighted image to the diffusion data, further spatially smoothed using a Gaussian kernel with a standard deviation of 3 mm to introduce a smooth boundary extending slightly beyond – as well as within – the edges of the tumor.
For scientific reference, that’s this source (Aerts et al., 2019): Evaluating the performance of 3-tissue constrained spherical deconvolution pipelines for within-tumor tractography. (https://doi.org/10.1101/629873)
@Hannelore_Aerts and myself developed this solution. Hannelore left active research upon finishing her PhD, so we didn’t get round to get this fully out at the time. The same observations will make it into another published work though; no worries.
In any case, I can directly help you with the approach if you wish. It requires some careful QA in the process. From a first look at your screenshots and the description you provide, it should be suitable in your specific setting. Just drop me an email if you’re interested.
Cheers,
Thijs

