Hello,
I posted a question about registering my tractogram to MNI152 space a while ago (Registration of tractogram to MNI152 space) and am now encountering new difficulties. I followed the instructions posted last year in this chat, however, I noticed that even though the normalization seems to work fine for the most part, there are some parts, where the tractogram partially lies outside of the MNI152 space.
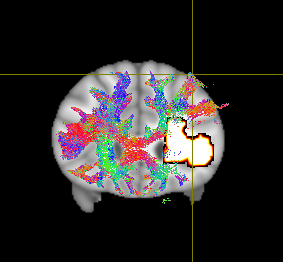
The code I used is the following:
#Register T1 (previously registered to DWI) to MNI152
flirt -ref MNI152_T1_1mm_brain.nii.gz -in T1_to_b0mean_FSLbbr.nii.gz -omat T12MNI_flirt.mat -out T12MNI_flirt.nii.gz
fnirt --ref=MNI152_T1_1mm_brain.nii.gz --in=T1_to_b0mean_FSLbbr.nii.gz --aff=T12MNI_flirt.mat --cout=warps_T12MNI --iout=T12MNI_fnirt.nii.gz
#Register MNI152 to T1 (previously registered to DWI)
invwarp --ref=T1_to_b0mean_FSLbbr.nii.gz --warp=warps_T12MNI --out=warps_MNI2T1
#initialize warp
warpinit MNI152_T1_1mm_brain.nii.gz inv_identity_warp_no.nii.gz
#Apply transformation to identiy warp
applywarp --ref=b0_upsaml_bet.nii.gz --in=inv_identity_warp_no.nii.gz --warp=warps_MNI2T1.nii.gz --out=mrtrix_warp_MNI2dwi.nii.gz
#Transform MNI152 to DWI
mrtransform MNI152_T1_1mm_brain.nii.gz -warp mrtrix_warp_MNI2dwi.nii.gz MNI2dwi.nii.gz
#Transform .tck file
tcktransform ifod2_5M.tck mrtrix_warp_MNI2dwi.nii.gz tck2MNI.tck
In the code I am using the inverse transformation matrix of T1/DWI to MNI152, however, would it not be better to use the forward transformation from T1/DWI to MNI152?
Do you have any ideas what the issue could be?
Thank you,
Franziska