Dear MRtrix3 Team,
Hello, I hope this post finds you well.
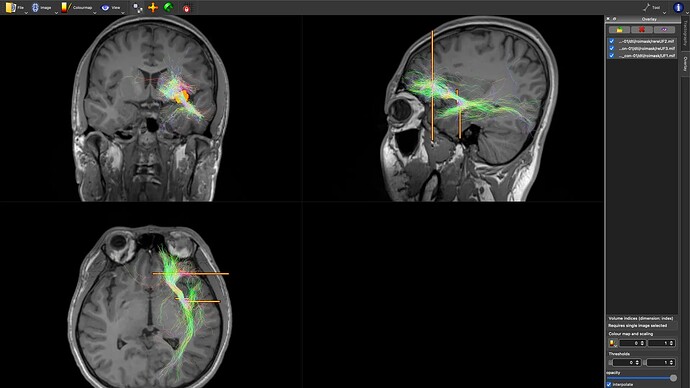
I generated a probabilistic (iFOD2) tractogram, as shown in the image below:
While it appears valid, many responses I’ve seen in the community suggest that a simple visual inspection cannot guarantee the reliability of a tractogram.
To address this, I decided to validate the pipeline I used by examining large tracts using manually drawn landmark ROI masks. I specifically checked the UF, ILF, IFOF, and CC tracts.
-
UF:
-
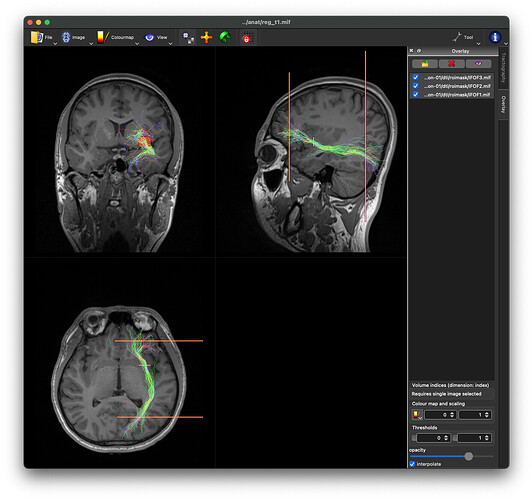
ILF:
-
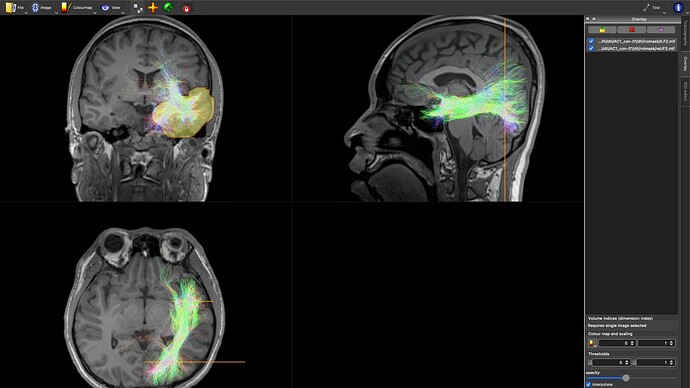
IFOF:
However, the results do not seem to align with reliable cases I have observed. In my opinion:
- The UF and ILF tracts are somewhat similar to what they should look like.
- The IFOF tract is entirely incorrect.
- The CC appears overly dense.
I am now unsure whether my pipeline is correct. I would like to ask for your guidance:
- I don’t think there’s a definitive answer, but could you at least help me judge whether the tractogram I created looks odd or not?
- If the tractograms are indeed problematic, could you help identify which part of my preprocessing pipeline might be causing the issue?
I have attached my preprocessing and tractography generation code below for your reference.
export PATH="/usr/local/mrtrix3/bin:$PATH"
bpath="/Volumes/research/source/nifti/dti/"
spath="/Volumes/research/source/nifti/surf/surf/"
cpath="/Volumes/research/code"
export FREESURFER_HOME=/Applications/freesurfer/7.3.2/
source $FREESURFER_HOME/SetUpFreeSurfer.sh
export SUBJECTS_DIR="${spath}"
atlas="atlasA"
atlas_name="atlA"
export nstr=1M
export tckfname=tck_ACT_seedmask_${nstr}
export nthr=10
cd ${bpath}
for csub in $(ls -d A*); do
cd ${bpath}/${csub}/dti/
export dwiap=${bpath}/${csub}/dti/${csub}_dwi_ap.mif
export dwipa=${bpath}/${csub}/dti/${csub}_dwi_pa.mif
dwi2mask ${dwiap} mask.mif
maskfilter mask.mif dilate dmask.mif -npass 3
dwidenoise ${dwiap} denoise.mif -noise noiselevel.mif -mask dmask.mif
mrdegibbs denoise.mif degibbs.mif
dwiextract -bzero ${dwiap} - | mrconvert - b0_ap.mif -coord 3 1,2,3,4,5,6,7,8 -force
dwiextract -bzero ${dwipa} b0_pa.mif
mrcat b0_ap.mif b0_pa.mif b0pair.mif
dwifslpreproc ${dwiap} geomcorr.mif -rpe_pair -se_epi b0pair.mif -pe_dir ap -readout_time 0.0383 -align_seepi
dwibiascorrect ants geomcorr.mif biascorr.mif -bias biasfield.mif -force
dwi2mask geomcorr.mif mask_geomcorr.mif -force
maskfilter mask_geomcorr.mif dilate dmask_geomcorr.mif -npass 3 -force
dwi2tensor -mask mask_geomcorr.mif biascorr.mif dt.mif -force
tensor2metric dt.mif -fa dt_fa.mif -ad dt_ad.mif -rd dt_rd.mif -adc dt_adc.mif -vector dt_ev.mif -vec dec.mif -force
dwiextract biascorr.mif -shells 1000 - | mrcalc - $(dwiextract biascorr.mif -bzero - | mrmath - mean -axis 3 -) -div norm_b1000.mif -force
dwi2response tournier -mask mask_geomcorr.mif norm_b1000.mif rf_norm_wm_trn.txt -force
tail -1 rf_norm_wm_trn.txt > rf_norm_wm_trn_tail.txt
dwi2fod csd norm_b1000.mif -mask mask_geomcorr.mif rf_norm_wm_trn_tail.txt norm_wm_fod_trn.mif -force
dwiextract biascorr.mif -bzero - | mrmath -axis 3 - mean - | mrconvert - mean_b0_biascorr.nii -force
flirt -dof 6 -cost normmi -in ../anat/${csub}_t1w.nii -ref mean_b0_biascorr.nii -omat ../anat/T_fsl.txt
transformconvert ../anat/T_fsl.txt ../anat/${csub}_t1w.nii mean_b0_biascorr.nii flirt_import ../anat/T_T1toDWI.txt -force
mrtransform -linear ../anat/T_T1toDWI.txt ../anat/${csub}_t1w.nii ../anat/reg_t1.mif -force
mrconvert ../anat/reg_t1.mif ../anat/reg_t1.nii
bet2 ../anat/reg_t1.nii ../anat/reg_t1_bet -m -f 0.4
gzip -d ../anat/reg_t1_bet.nii.gz
gzip -d ../anat/reg_t1_bet_mask.nii.gz
mrconvert ../anat/reg_t1_bet.nii ../anat/reg_t1_bet.mif -force
mrconvert ../anat/reg_t1_bet_mask.nii ../anat/reg_t1_bet_mask.mif -force
5ttgen fsl ../anat/reg_t1.mif ../anat/5ttseg.mif -nocrop -force
5tt2vis ../anat/5ttseg.mif ../anat/5ttvis.mif -force
mrconvert ../anat/5ttvis.mif ../anat/5ttvis.nii -force
5tt2gmwmi ../anat/5ttseg.mif ../anat/gm_seed.mif
mri_aparc2aseg --s ${csub}_t1w.nii --annot ${atlas} --new-ribbon --annot-table $FREESURFER_HOME/renum_${atlas}.txt --noribbon
mrconvert ${spath}/${csub}_t1w.nii/mri/${atlas}+aseg.mgz ${bpath}/${csub}/anat/${atlas}+aseg.nii
mrtransform -interp nearest -linear ../anat/T_T1toDWI.txt ../anat/${atlas}+aseg.nii ../anat/reg_${atlas}+aseg.nii -force
matlab -batch "addpath ('${cpath}'); renum_atlas('${bpath}/${csub}/anat/reg_${atlas}+aseg.nii')"
mrconvert ../anat/reg_${atlas}+aseg_conse.nii ../anat/reg_${atlas}+aseg_conse.mif
tckgen norm_wm_fod_trn.mif ${tckfname}.tck -select ${nstr} -seed_image ../anat/gm_seed.mif -act ../anat/5ttseg.mif -backtrack -crop_at_gmwmi -maxlength 250 -angle 60 -cutoff 0.06 -info
tcksift2 ${tckfname}.tck norm_wm_fod_trn.mif tck_weights_${tckfname}.txt -act ../anat/5ttseg.mif -out_mu SIFT2_mu_${tckfname}.txt -out_coeffs tck_coeffs_${tckfname}.txt
tck2connectome ${tckfname}.tck ../anat/reg_${atlas}+aseg_conse.mif conn_${tckfname}_${atlas_name}.csv -tck_weights_in tck_weights_${tckfname}.txt -out_assignments assign_${tckfname}_${atlas_name}.txt
echo "Processing of $csub completed."
done
Thank you very much for your time and assistance. I look forward to your feedback and any advice you might have.
Best regards,
Narae Kim