Hi Donald,
Thanks for your reply. Bear with me as I try to explain what we know so far.
I agree that the directions defined in philips can be confusing because they depend on the scan setting (e.g. gradient overplus, input file, or mps file, etc). In our case, I’m certain that all the directions (opt12 and directions in DICOM) in our scans before and after August 2021 were consistent. Our scan protocols were consistent: We inspected the exam cards. We used classic dicoms. The private tags were consistent: (2005,1563) = sagittal slice order = LR, (2005,1564) = coronalsliceorder = PA and (2005,1565) = transversalsliceorder = FH. This suggests our MPS directions were in RAS+; The transformations from opt12 (with respect to image axes) to the directions in DICOM (LPS+ = with respect to the patient-centered coordinate system) were consistent: I figured the rotation sequence from the investigation scan and applied it on our previous angulated scans. In mrtrix, the dw_scheme was in RAS+ (with x and y axes flipped); The bvecs/bvals in LAS+ were consistent: All our participants gave the same bvecs/bvals (with the x-axis flipped).
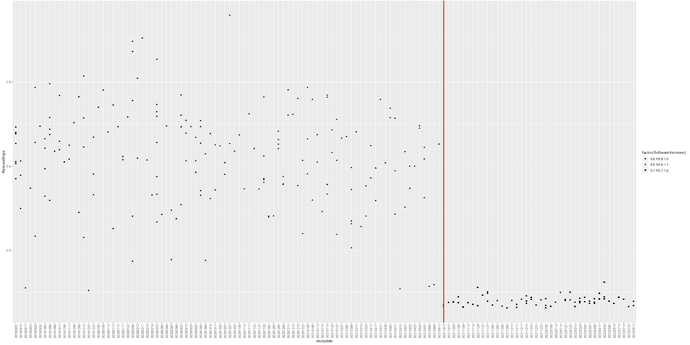
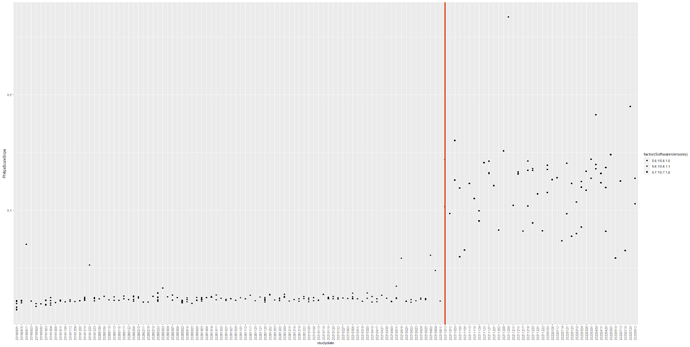
Then, I started comparing the DICOM headers to see if they were the same. 3 things were inconsistent. First, the number of fields in the DICOM changed after the software updated. Second, the rescaleslopetag (0028,1053) were inconsistent. Third, the philipsscaleslope tag (2005,100E) were inconsistent. See the timeline below. Could this trigger something? This suggest that the dicoms before and after August 2021 were inconsistent despite the same scan protocol.
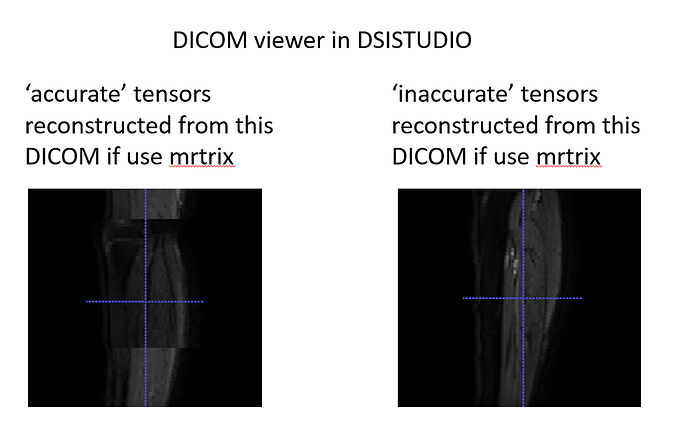
Another related finding is that the dicom viewer of the dsistudio sees the dicoms before and after August 2021 differently, although we did not find any obvious issue in mrtrix as well as dcm2niix (by Chris Rorden). Its confusing here because dsistudio sees our more ‘accurate’ scans wrongly. Anyway, this suggests that the dicoms may have been stored differently. Earlier, mrtrix had slice sorting issue but this has been resolved in the latest release: Problem with sorting of DICOM images using mrconvert (thanks for that!)
If you find that running a direct DICOM → tensor → eigenvector pipeline all within MRtrix produces inconsistent results, then we’ll need to investigate whether that’s due to a bug in our DICOM handling or in the latest version of the Philips software… Try a command such as this one to do this:
With regards to your suggestion to reconstruct primary eigenvectors directly from the DICOMs, here are the results:
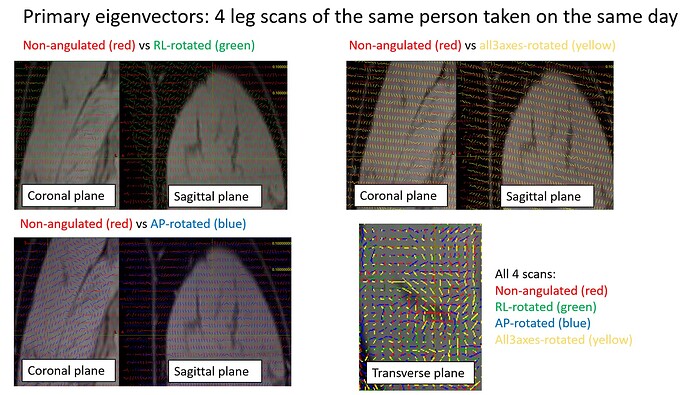
To recap, we conducted an investigation study comprised of 4 DTI scans: one not-angulated scan (ie with the image axes aligned with the scanner), one with a +20 degr rotation around the RL axis, one with +20 degr rotation around the AP axis, and one with rotations applied in all 3 planes (which is how we typically obtain for our scans): +7degr around RL axis, -2.84 degr around AP axis and +16.5 degr around IS (or philips calls this PH) axis. We exported classic dicoms, par/recs, enhanced dicoms and xml/recs.
Tensors and tracts from the PAR/RECs and ENHANCED DICOMs appeared consistent with those from CLASSIC DICOMs. However, one thing worth knowing is that the private tags (0018,9075) = Diffusion Directionality in all the files in CLASSIC DICOM were shown as ‘NONE’ whereas these tags were different in the files in ENHANCED DICOMS (ie it was shown as ‘NONE’ for the b0 slices and ‘DIRECTIONAL’ for other slices containing gradient directions).
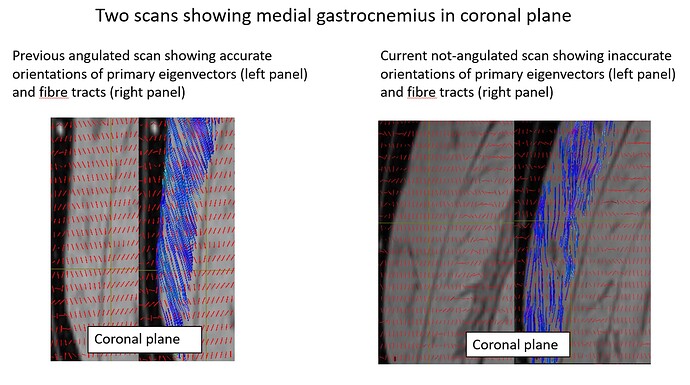
Below, I show one scan (on the left column) conducted before August 2021. Typically, we would expect to see the fibre tracts with respect to the aponeurosis (shown as dark voxels) to be angled roughly this way. If you compare it with the not-angulated scan, you can see that the tracts of the not-angulated scan were somewhat straighter. So even the not-angulated scan was already giving us a problem.
Here, I compare primary eigenvectors between the 4 scans. You can see very clear differences in orientations despite they were scanned on the same person and on the same day.
FYI, our engineers ran some QC checks on the hardware including the coil but did not find anything unusual. We are hoping its a software bug though. Fingers crossed.
To summarise, we saw consistent directions between participants but there were inconsistencies in image scaling and DICOM handling. Could it be a software bug somewhere? We are happy to share our data if it helps. Please let us know.
Cheers, Brian