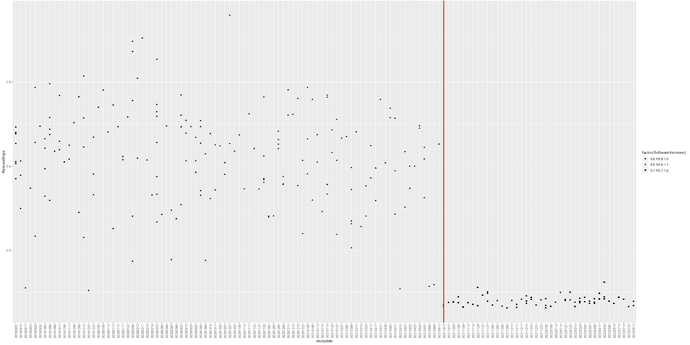
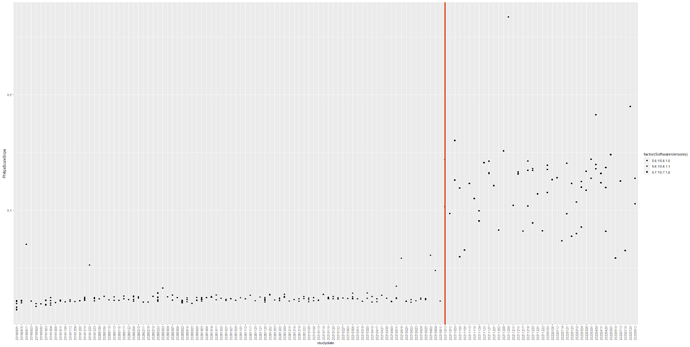
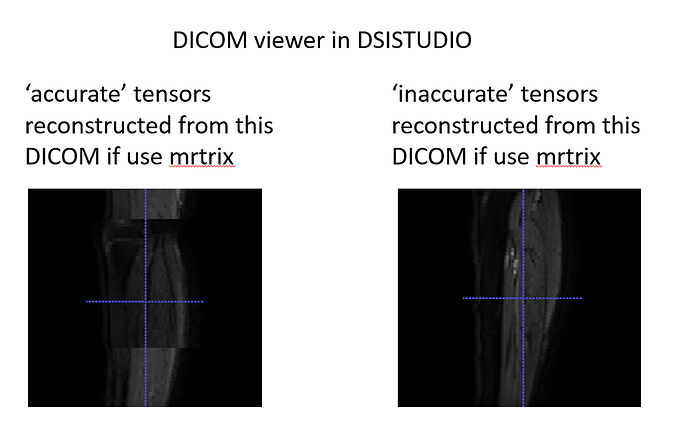
Hello. We used deterministic tractography on our DTI data (taken from a Philips scanner) to generate muscle fibres in the lower limbs. The fibre tracts of our first roughly 150 scans which were scanned before August 2021 were okay. We found that the orientation of the fibre tracts after August 2021 were implausible despite that the scan protocols between participants were the same. Did anyone had this issue before? We don’t really know why at this stage, was hoping someone has faced this issue before.
We noticed that the Philips software was updated from 5.6.1\5.6.1.1 to 5.7.1\5.7.1.2 in August 2021 (still not sure if this was the reason. we will need to investigate this). We are currently using the latest mrtrix version as shown below.
Here’s what we have investigated:
- We tried using both .mif and .nii+fslbvecs separately. Both methods were reproducible.
- We checked the raw gradient directions and the fsl bvecs between participants, before and after that date. The orientation of the raw gradients (with respect to the scanner coordinate system) between participants were different. The orientation of the fsl bvecs (with respect to the image axes in LAS coordinate system) between participants were the same.
- The x-axes of the fsl bvecs (in LAS format) for all the participants were being flipped when compared to the original vector scheme (which was in LPS format in patient coordinate system). Should we expect the y-axis being flipped instead?
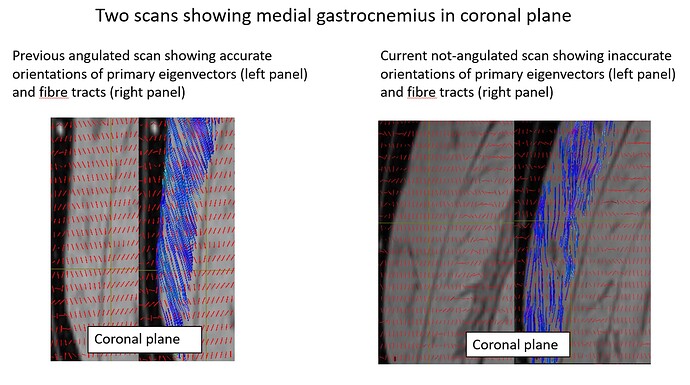
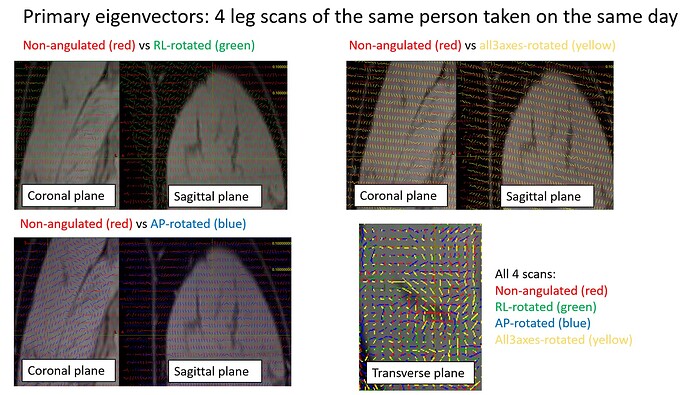
- We recently conducted separate investigation scans by using the same DTI protocol but with various stack angulations. We started with a scan with zero angulations in all the 3 planes (ie all 3 field of views being aligned with the scanner) and some other scans with one or all of the planes being rotated. All the scans including the aligned scan did not give plausible results. We also looked at the raw primary tensors before fsl topup (as in without any motion or eddy current corrections), they all looked very differently orientated, despite no obvious motions and the scans were taken on the same day.
- We were not able to use the dwgradcheck method because it was not designed to deal with muscle architecture.
- We checked the raw DTI scans between participants, there were no obvious artefacts.
Are we missing anything? Could someone suggests if there are other ways to troubleshoot this? Appreciate it.