Hello,
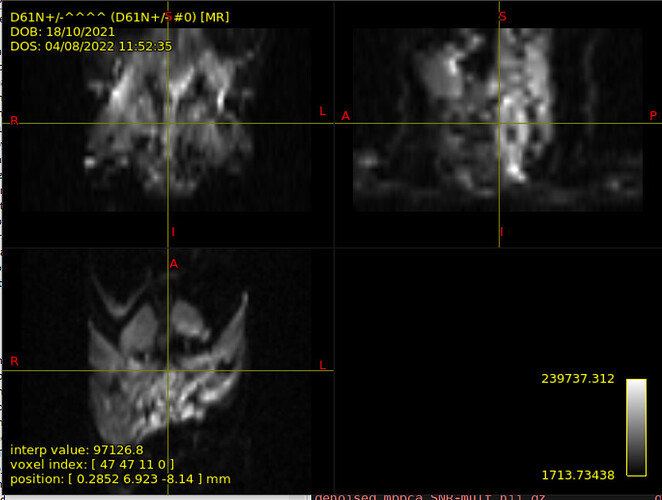
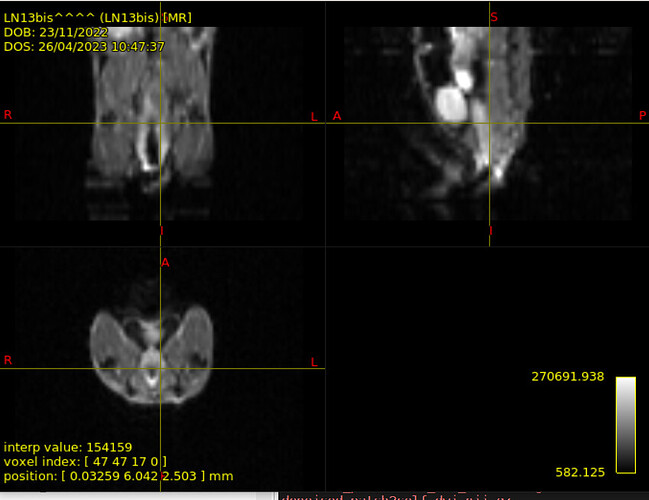
I have some mouse DTI data I am working with. The acquisition protocol is made up of 5 b=0 and 30 b=700 s/mm2 so it is strictly DTI. Moreover, they concern peripheral nervous system, since the target is the sciatic nerve – you have an example in the image attached. Some images have been acquired with surface coil, other with volume coil. Surface-coil one in particular are quite artefactual and I wanted denoised them via Patch2Self at first, to show the improvement gained in image quality thanks to denoising. I then extracted average DTI measures in certain ROIs corresponding to sciatic nerve in the slices where it’s visible. I then wanted to perform tractography to track the sciatic nerve. However, I do have T2 images but they are not coregistered to DTI ones…I guess they have been acquired at two different times. I was woondering whether I could perform tractography all the same without anatomy, and in case how.
Thanks in advance,
Rosella
→ volume DTI scan