Hi @sekara98,
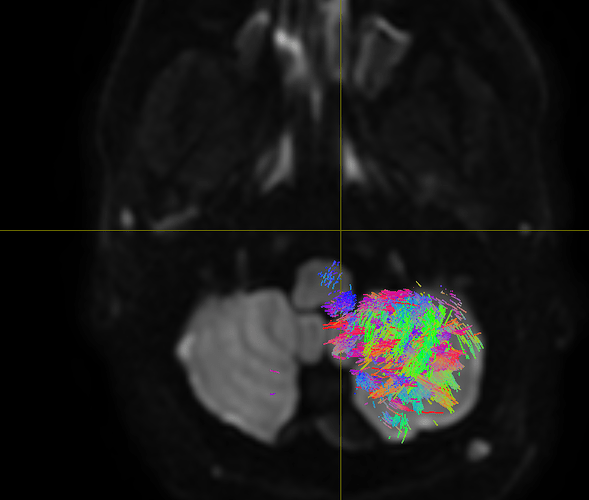
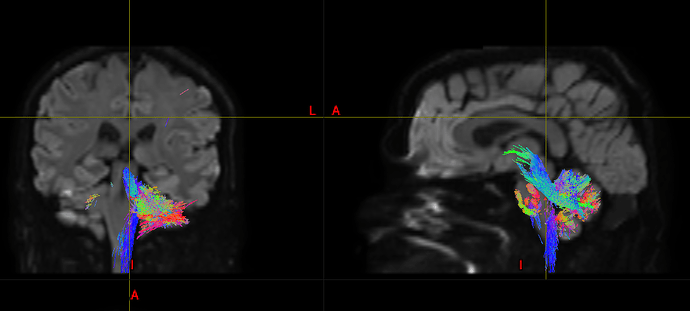
Performing tractography in the cerebellum is not an easy task, as the complexity of both white matter internal structure and grey matter folding is even greater than that of the cerebrum. So given it’s your first time doing tractography, from a student supervision perspective I would actually suggest commencing with doing some arbitrary experiments in the cerebrum, getting a feel for the technology and the software, and only then starting to look at the cerebellum and the additional challenges that it presents.
All the images have been pre-processed and 4 cerebellar ROIs (right/ left cortex, right/left white matter) have been segmented using the Freesurfer labels.
I would firstly note that the FreeSurfer cerebellar labels are quite coarse. The “grey matter” labels essentially capture not only the grey matter but also the deepest white matter projections as kind of large amorphous blobs. I’ve had some success with using just a basic image-intensity-based segmentation in just the cerebellum to get a little more GM-WM detail.
tckgen -select 0 -seeds 250000 -cutoff 0.15 -angle 6 -seed_image label_7.nii -seed_unidirectional fod.mif label_7.tck -force
The primary problem here is “-angle 6”. What this means is that for every half-voxel distance, streamlines are not permitted to curve more than 6 degrees. That’s why all of your streamlines look almost straight. The default is 45 degrees. Your FOD amplitude cutoff is also much higher than the default, which actually also makes streamlines tend to be straighter than otherwise (though this takes a bit more space to explain).
Secondly I wanted to know how to select the number of seeds and the different parameters?
Well, that’s a bit of a how-long-is-a-piece-of-string question. For number of seeds / streamlines, I’d echo an argument I’ve made in many threads (and will try to better formalise in print at some point), which is that if you re-generate your streamlines, redo the rest of your analysis, and it yields a different result, you didn’t generate enough streamlines in either case. This is an experiment-specific question, so it’s worth actually doing. I’m also generally suspicious of anything supposedly quantitative where I can make out visually individual streamlines, as it implies that the total space of all possible streamline trajectories has not been sampled adequately densely.
For the second part I have seen the use of different atlases but am unsure how to about this.
The fundamental process should be no different to that of studies of the cerebrum, though it’s possible that targeted registration of just the cerebellar volume may benefit spatial alignment. There’s a Wiki page here, and see also the BATMAN tutorial.
Cheers
Rob