Good evening MRTrix3 Experts,
I am running CSD probabilistic tractography on the S900 release of the Human Connectome Project dataset. I am trying to convert my .tck files to MNI space, and to do this I followed @eloydelas instructions from 2016, which can be reviewed at this link. My ultimate goal is to do the following:
- Transform .tck tractographies to MNI space
- Convert tractographies to voxel maps
- Average together tractographies for all subjects in a common space to create a spatial histogram/probabilistic map using a function like fslmaths in FSL.
- Calculate Dice Coefficients for the resultant map to assess overlap with ROIs from various atlases
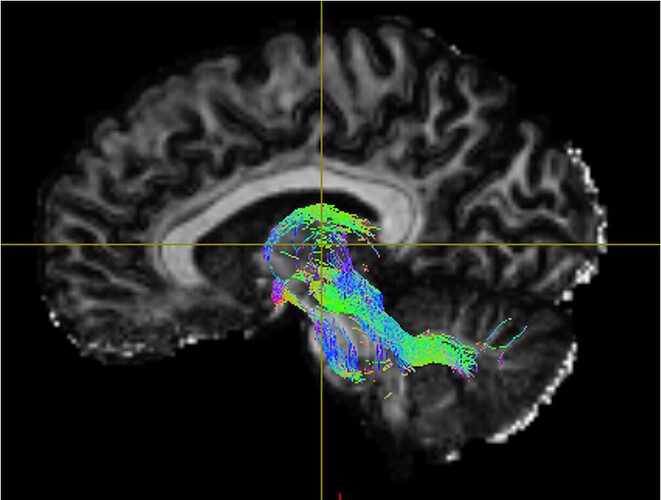
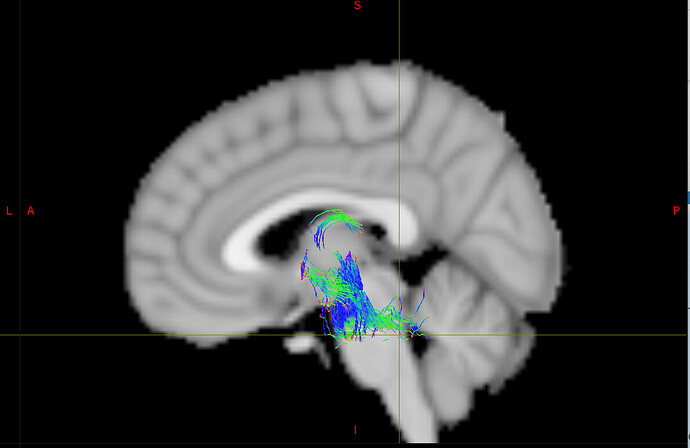
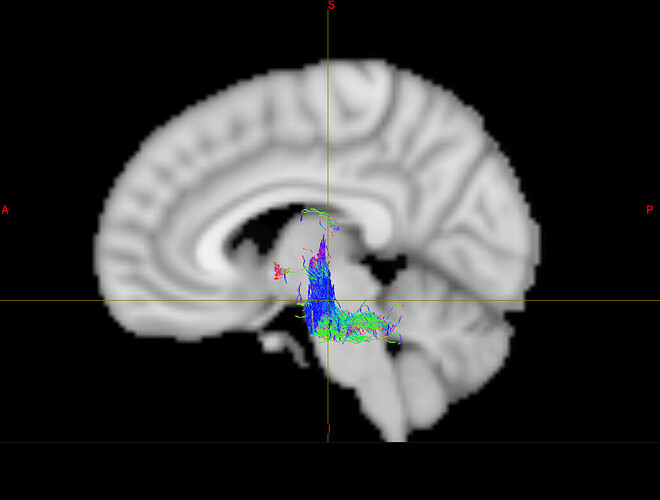
- Below are examples of subject-space tractographies, and my failed MNI normalization of the same tract:
Subject-Space Tractography:
Failed MNI Normalisation:
My pipleline is delineated as follows:
- Create no-warp images in target space:**
warpinit \
$FSLDIR/data/standard/MNI152_T1_2mm_brain.nii.gz \
no-warp-[].nii
- Warp target images to subject space:
for s in ${subjects}
do
for n in 0 1 2
do
fnirt \
--ref=dtifit_output/DTI_FA.nii.gz \
--in=no-warp-${n}.nii \
--aff=standard2diff_${s}.mat \
--iout=warp-${n}_${s}.nii
done
done
- Perform MNI normalisation on .tck images:
for s in ${subjects}
do
tcktransform \
ifod2_paths.tck \
warp-[]_${s}.nii \
ifod2_paths_mni.tck
done
Please note that I also tried redoing this process perfoming the following intermediary step in FSL:
for s in ${subjects}
do
for n in 0 1 2
do
fslorient \
-copysform2qform \
warp-${n}_${s}.nii
done
I would sincerely appreciate any advice this community may be able to offer!
Kind regards,
Linda