So here’s what that looks like plotted in 3D:
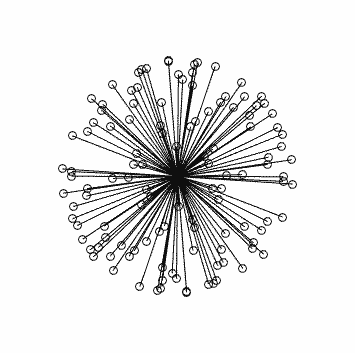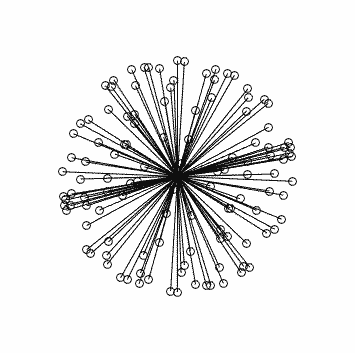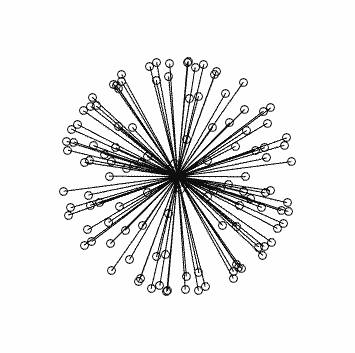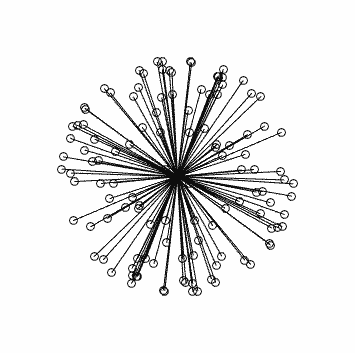
Clearly this is very far from optimal…
This is what you should see (generated using dirgen 60 dir60.txt -cartesian:
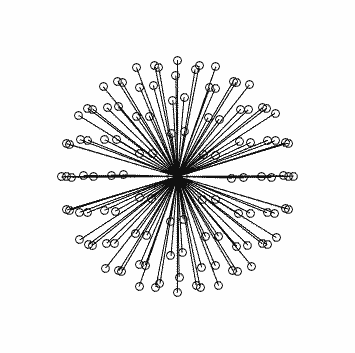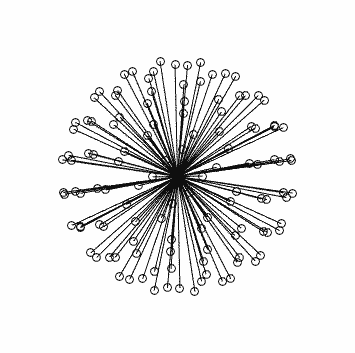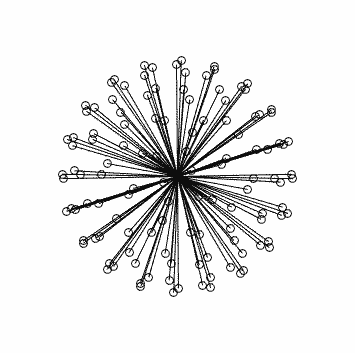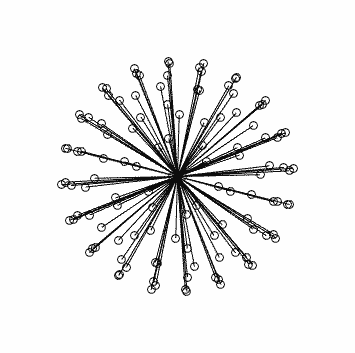
with corresponding stats in this case:
$ dirstat dir60.txt
dir60.txt [ 60 directions ]
Bipolar electrostatic repulsion model:
nearest-neighbour angles: mean = 18.9073, range [ 18.114 - 19.9099 ]
energy: total = 8002.93, mean = 133.382, range [ 131.922 - 135.419 ]
Unipolar electrostatic repulsion model:
nearest-neighbour angles: mean = 19.3059, range [ 18.1859 - 31.9532 ]
energy: total = 3999.4, mean = 66.6566, range [ 40.5186 - 82.3908 ]
Spherical Harmonic fit:
condition numbers for lmax = 2 -> 8: 1.00556 1.02185 1.05982 1.20328
Note that the worst condition number for the SH fit is 1.2 at lmax=8, compared to your 12.7592…
So I’m not sure where your directions came from, maybe they were somehow modified along the way? But they’re clearly nowhere near optimally distributed…
No, gen_scheme.sh uses dirgen internally to generate the directions for each shell. I recommend you keep using that script, but figure out how to massage the output into a format suitable for Siemens. Something like this should do it:
bmax=3000
cat dw_scheme.txt | awk '{ n=sqrt($4/'$bmax'); printf "Vector["NR"] = ( "; if (n>0) printf $1*n ", " $2*n ", " $3*n " )\n"; else printf "0, 0, 0 )\n"; }'
This will scale the vectors by sqrt(b/bmax), which is what I think is the right scaling (more on that later), and produce them in a format that can be directly inserted into your DiffusionVectors.txt file. You may need to amend it to fit your needs though, and you’ll also need to insert something like this at the top of the output (again, make sure it fits with what you’ve got):
# multi-shell set, generated using gen_scheme
[directions=106]
CoordinateSystem = xyz
Normalisation = none
So about getting the scaling right for this to work, I note you quoted:
Where does this come from? I’d assume that the right scaling here should actually be sqrt(1000/3000) = 0.57735, not 0.33 - this is what the code in the MRtrix3 backend assumes, and I’m pretty sure we verified this was correct at the time…
That would be a phenomenal waste of resources: this is standard resolution at 1.5T… ![]()
No, you should be able to push the resolution up to maybe 1.8mm isotropic without much of an issue, it’s just that as you lower that, your SNR will drop rapidly, so you’ll need to figure out what works best. You should aim for an SNR in the b=0 image no lower than 15 - bearing in mind that SNR will be strongly spatially variable at 7T, so you’ll need to measure it from the worst location SNR-wise that you care about (most likely the brainstem, in my experience).
Yes, that’s what it’s supposed to do.
Yes, that would definitely be a problem for MSMT-CSD. You can try to scale each acquisition so that the b=0 image intensities match, as long as none of the other acquisition parameters differ between the acquisitions. You can use mrcalc for that, in combination with mrstats to get the b=0 intensities. dwiextract -bzero may also come in handy here. I’d also recommend sticking to within a brain mask to avoid undue influence from the background noise here. This might look something like:
# generate a brain mask (use the same mask for all acquisitions):
dwi2mask dwi.mif mask.mif
# then for each acquisition, get mean b=0 image intensity within mask:
sig=$(dwiextract dwi.mif -bzero - | mrmath - mean -axis 3 - | mrstats - -mask mask.mif -output mean)
# and scale to have mean b=0 signal = 1000:
mrcalc dwi.mif 1000 $sig -div -mult dwi_scaled.mif
# once you've done that for all of them, recombine with mrcat:
mrcat dwi_scaled1.mif dwi_scaled2.mif dwi_scaled3.mif -axis 3 dwi_all_scaled.mif
This is just rough and ready, you’ll no doubt need to amend to suit your data, but that should give you an idea of what you can do…
No, you’re correct. As long as you can provide a DifffusionVectors.txt file to the system in the right format (as above), you should be fine. What @ThijsDhollander what referring to was that this relies on you having the right license in place - it’s not enabled unless you’ve purchased the appropriate diffusion package…