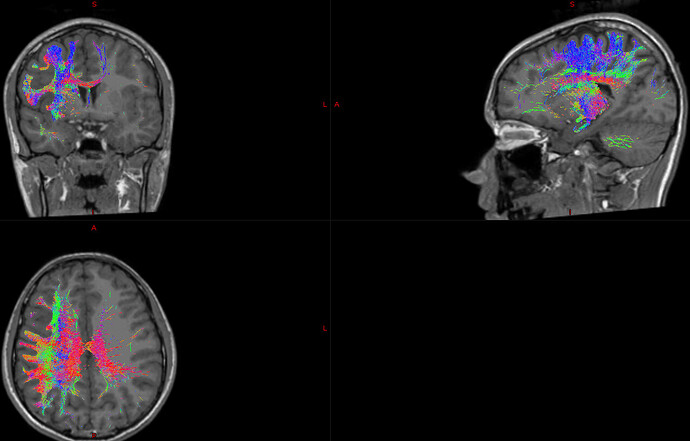
After processing all the different masks necessary for the representation of the different main tracts found in the CNS, I’ve got this kind of images:
I can’t understand why there is a right hemisphery tract on my image if I told, specifically, tckgen to exclude all the information found on that hemisphere. The code used is the next:
tckgen -algorithm iFOD2 -fslgrad …/BThings/Suj_28.bvec …/BThings/Suj_28.bval -act annotations/Suj_28/registration/Suj_28_5TT_b0space.nii.gz -seed_image annotations/Suj_28/registration/Suj_28_AAL_rh_slfp_mask.nii.gz -select 1M -maxlength 250 -cutoff 0.06 -crop_at_gmwmi -include …/Freesurfer/Suj_28_FreeSurfer/dlabel/mask/rh_slfp_b0_mask.nii -exclude results/lh_AAL.nii results/Suj_28_structural_measures/Suj_28_wm_fod_norm.nii.gz results/Suj_28_structural_measures/Suj_28_1M_seeds_AAL_slfp.tck
the image has been edited with tckedit
tckedit results/Suj_28_structural_measures/Suj_28_1M_seeds_AAL_slfp.tck results/Suj_28_structural_measures/Suj_28_100k_seed_AAL_slfp.tck -number 100k
Does anybody know what is going on?
Thanks in advance!