Hi all!
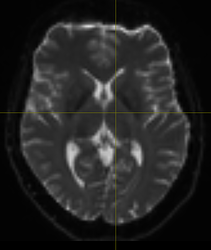
I’m working with a dataset with AP as the main phase encoding direction, and I also have the reverse PE acquisition for some b0 volumes. The thing is that the AP dwi present the typical warping artefacts and the very frontal region of the brain is missed:
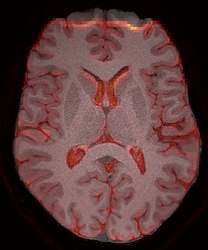
Caption: LEFT: mean b0 with AP. RIGHT: T1w image and the mean b0 overlaid (red).
During the preprocessing steps I executed:
dwifslpreproc dwiPA_denoise_unring.mif dwi_denoise_unring_preproc.mif -pe_dir PA -rpe_pair -se_epi b0_pair.mif -eddy_options "--slm=linear --data_is_shelled"
The output looks the same.
Is there any way to correct this artefact? Any thoughts?
One of the consequences is that, for example, the frontal pole area of the Desikan-Killiany atlas is missed in the dwi.
Thank you!
Stella