Dear Community,
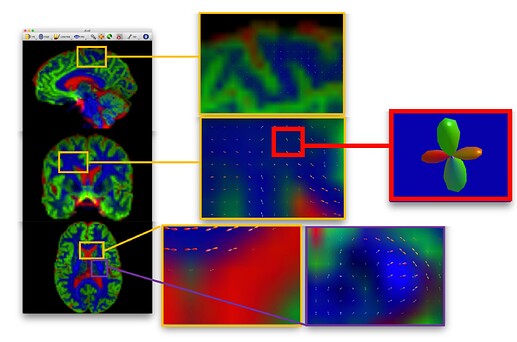
I’m generating FOD for my two shells(0,1500), 60 directions data, using the MSMT-CSD method.I want to check my generated fod map to make sure everything is ok. But I suddenly realised that there are very few tutorials on what the results of two shell analysis using MSMT-CSD should look like.The tutorial on Andy’s book and BATMAN, which used data from larger than two shells, obtained results similar to Figure 1.But I find this very different from the results I generated. Anyway I want to make sure that my results are normal. If it is wrong, is there something wrong there? I hope to get replies from all of you. Thanks.
This is the code I used to generate the WM,GM, CSF response function, and FOD.
# use dhollander
dwi2response dhollander preproc_unbiased.mif \
wm_response.txt gm_response.txt csf_response.txt -voxels voxels.mif
# Upsample
mrgrid preproc_unbiased.mif regrid -vox 1.25 preproc_unbiased_upsample.mif
# FOD MSMT,only WM and CSF
dwi2fod -force msmt_csd preproc_unbiased_upsample.mif \
-mask mask_upsample.mif \
wm_response.txt wmfod.mif \
csf_response.txt csffod.mif
# check result
mrconvert -coord 3 0 wmfod.mif - | mrcat csffod.mif - 2_tiusses_combin.mif
mrview 2_tiusses_combin.mif -odf.load_sh wmfod.mif
This is the result picture of BATMAN.
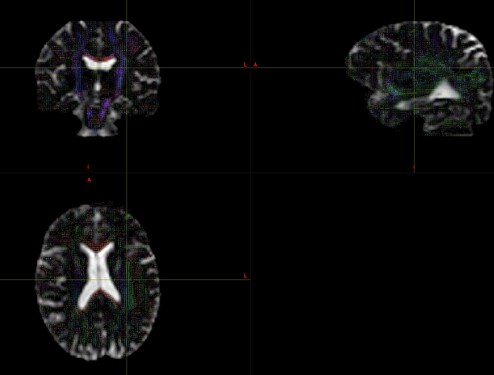
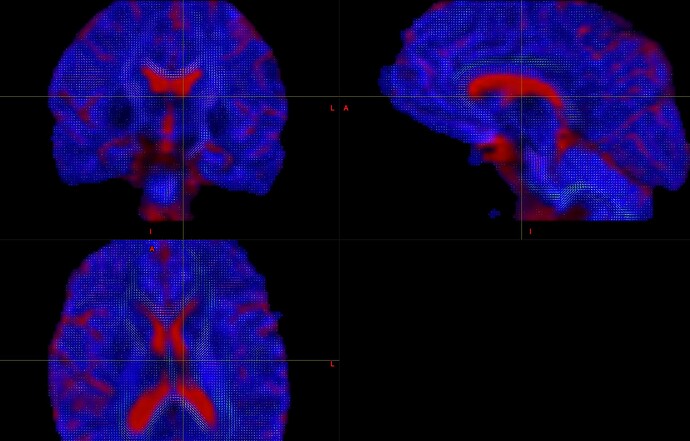
And,this is the picture of my result. I would like to know if this result is normal?
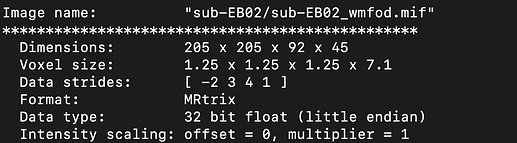
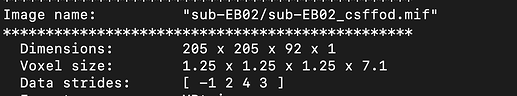
On top of that, I’m confused about the resulting wm_fod.mif and the cdf_fod.mif produced after dwi2fod is about? I checked both results using mrinfo and found that they have different volume counts. (The volume number of wm_fod is less than the volume number of my raw data,10b0 + 60 1500).
Also I noticed that ss3t_csd_beta1 is available on MRtrix3Tissue for single shell data. I wonder which method is right for my data?![]()
Anyway hope to get everyone’s input and if anyone has any insight please let me know. Thanks.
Yufeng