I’m trying to visualize the endpoints of streamlines that connect regions in a particular atlas. If I say:
`tck2connectome mytracks.tck myatlas.nii.gz -out_assignments myassignments.txt -assignment_end_voxels
cat myassignments.txt | awk ‘{w=0; if($1>0 && $2>0 && $1!=$2){w=1}; print w}’ > myweights.txt
tckmap mytracks.tck mytracks_conntdi.nii.gz -template myatlas.nii.gz -tck_weights_in myweights.txt -ends_only
`
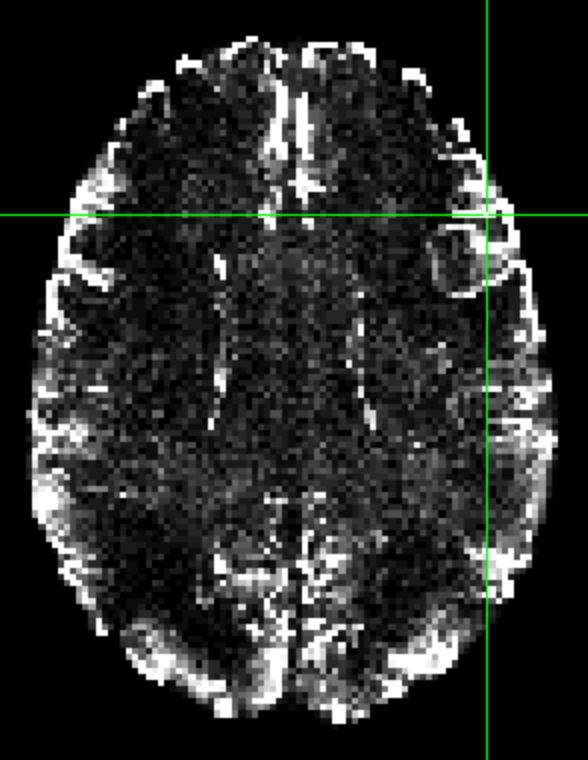
I get what I expect: non-zero counts in atlas voxels, and zeros outside the atlas voxels.
But if I do:
`tckresample -endpoints mytracks.tck mytracks_endpoints.tck
tck2connectome mytracks_endpoints.tck myatlas.nii.gz -out_assignments myassignments.txt -assignment_end_voxels
cat myassignments.txt | awk ‘{w=0; if($1>0 && $2>0 && $1!=$2){w=1}; print w}’ > myweights.txt
tckmap mytracks_endpoints.tck mytracks_endpoints_conntdi.nii.gz -template myatlas.nii.gz -tck_weights_in myweights.txt
`
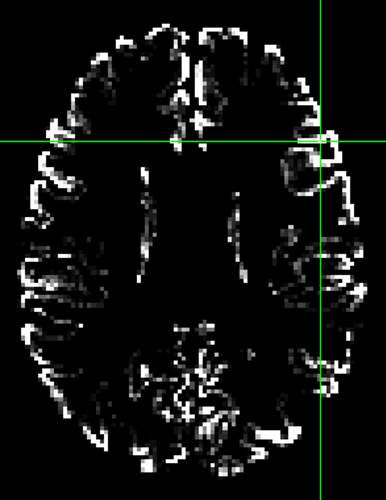
I get non-zero values well outside the atlas voxels. They are still generally much higher inside atlas voxels, but not exclusive to them, as i would expect.
I realize the weight file generation thing might be hackish, but aside from that, what is the difference between these two approaches?