Dear MRtrix community,
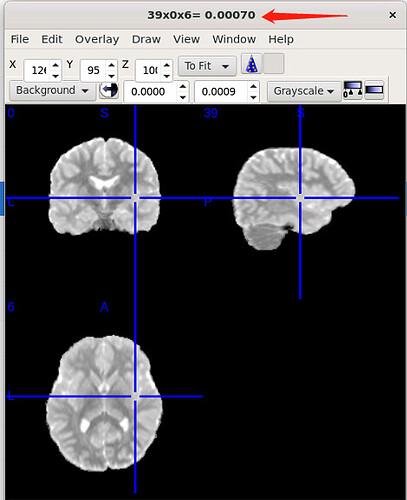
I used dwi2tensor and tensor2metric for the DTI fit. The generated MD.nii.gz, the value of the MD in the brain is around 0.0007, why is the order of magnitude so small? Is this normal for mean diffusivity value?
dwi2tensor dwi_denoise_preproc_bias.mif tensor.mif
tensor2metric -fa FA.nii.gz tensor.mif
tensor2metric -adc MD.nii.gz tensor.mif
tensor2metric -ad AD.nii.gz tensor.mif
tensor2metric -rd RD.nii.gz tensor.mif
Any help is greatly appreciated!
Panshi
Yes, these values are in the right range. The units for the diffusivities are mm²/s (opposite of the b-value, which is in s/mm²), and in the brain these are typically around 700-800 ×10-6 mm²/s – which matches what you show. Does that answer your question?
3 Likes
Thanks for your response, that was very clear and useful, as always!
Hi @jdtournier ,
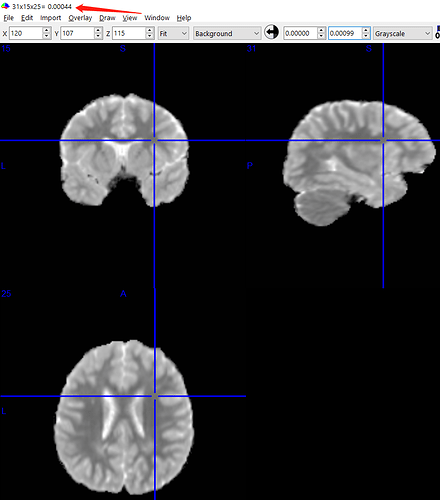
I find that my MD values are in the right range, but the numbers are still small. My MD values are about 500×10-6 mm²/s, while the normal one should be 670-800×10-6.
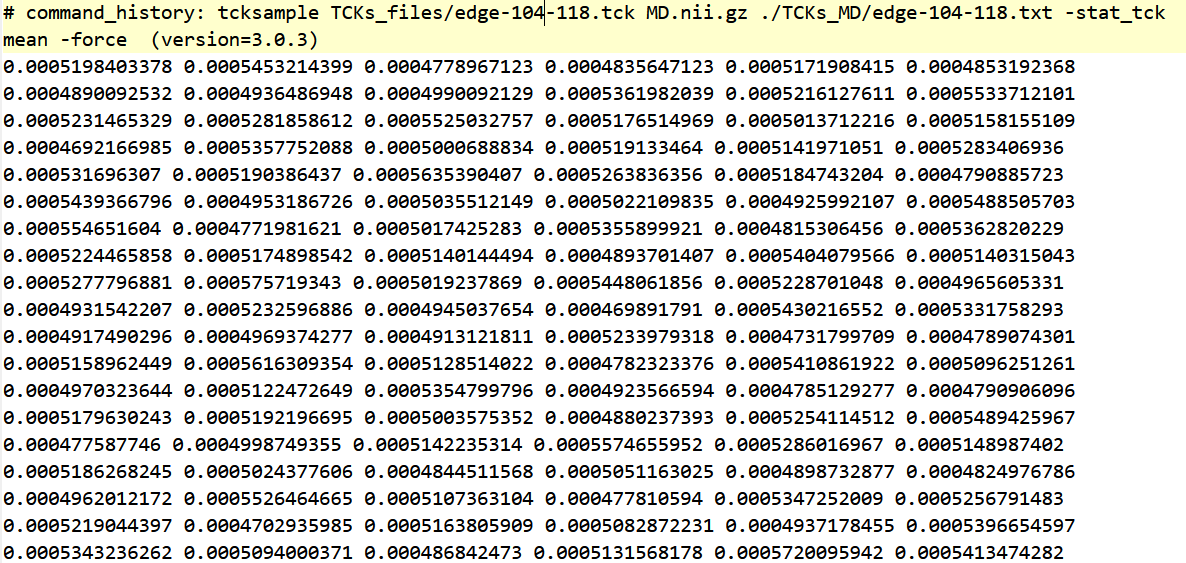
I used tcksample to get the MD value of a white matter edge and can see that the MD values of white matter are around 500 × 10-6 mm²/s. All subjects were like this.
My Code:
dwi2tensor dwi_denoise_preproc_bias.mif tensor.mif
tensor2metric -adc MD.nii.gz tensor.mif
tcksample TCKs_files/edge-104-118.tck MD.nii.gz ./TCKs_MD/edge-104-118.txt -stat_tck mean -force
I use the pre-processed ABCD DWI data, which is multishell.
In conclusion, the contrast of the MD map is normal (Grey and white matter are clear), but the overall value is small, as shown in the figure. What do you think are the possible reasons for the low MD values?
Thanks!
This is probably due to the high b-values in your sequence. At high b-values, the DTI model no longer provides a good fit because of the non-monoexpontential signal decay.
Instead of:
dwi2tensor dwi_denoise_preproc_bias.mif tensor.mif
try fitting the tensor only to the low b-values:
dwiextract dwi_denoise_preproc_bias.mif -shells 0,500,1000 - | dwi2tensor - tensor.mif
2 Likes
Thank you! @bjeurissen. You have answered my question.
The MD values I fitted using the three shells 0,500,1000 are indeed in the normal range. I should use all shells for CSD tractography and use inner shell for DTI fitting.
Also, why is the contrast of the MD map fitted with the three shells 0,500,1000 not as good as the one with the full shell?