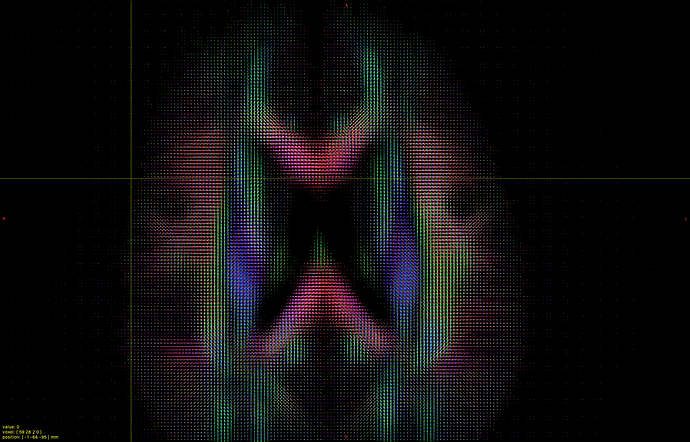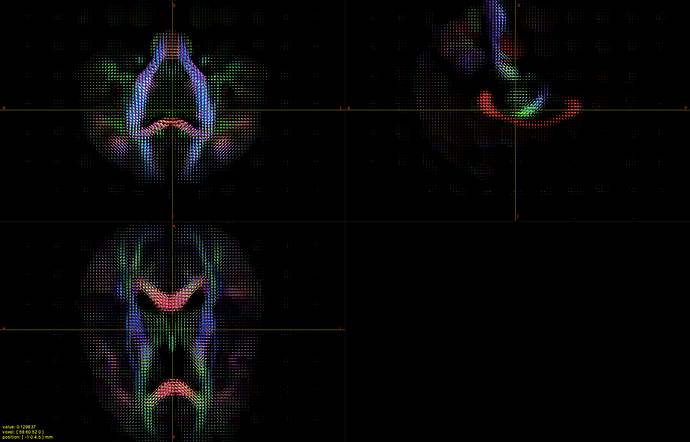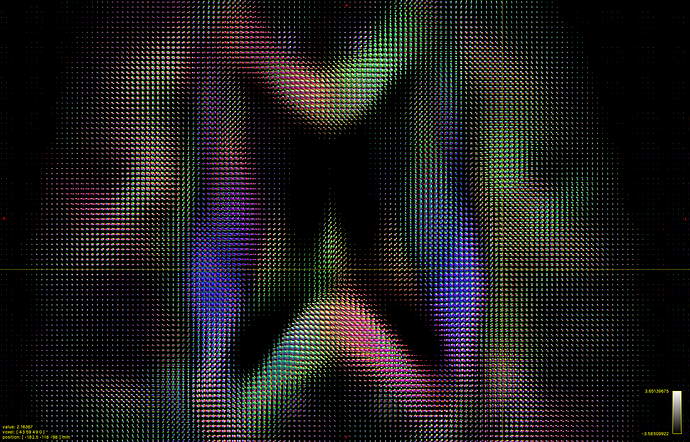Dear Tournier,
c= [0.10931488, -0.01913602, -0.10427029, 0.07533529, 0.02588572,
-0.06559721, 0.01725084, 0.03923653, -0.02286121, -0.04493933,
-0.00923942, 0.00201445, -0.07635243, -0.04356742, 0.02039322,
-0.00857603, -0.01146724, 0.01803261, 0.01759768, -0.00576732,
-0.01670703, 0.00723215, -0.0077153 , -0.00871654, -0.02096124,
0.02479116, 0.02442713, -0.00403376, 0.0023532 , 0.00225714,
-0.00634937, -0.00323842, 0.00272784, 0.00499127, -0.00106389,
-0.0041335 , 0.00481908, -0.0025849 , 0.00145664, -0.00031923,
0.00514136, 0.00942071, -0.00342335, -0.00609509, -0.00026663];
cc=reshape(c',[1,45]);
dx = pi/60;
% dx = pi/30;
col = 0:dx:pi;
az = 0:dx:2*pi;
[Phi,Theta] = meshgrid(az,col);
for l = 0:2:8
p = legendre(l,cos(Theta)');
for m = -l:l
clear v
j = (l^2 + l + 2)/2 + m;
if m<0
c1 = factorial(l-m)/factorial(l+m);
c2 = (2*l+1)/(4*pi);
c3 = (-1)^(abs(m))*(factorial(l-abs(m))/factorial(l+abs(m)));
v=reshape(p(abs(m)+1,:),[size(exp(1i*m*Phi),1),max(size(p(abs(m)+1,:),1),size(p(abs(m)+1,:),2))/size(exp(1i*m*Phi),1)])
Ylm = sqrt(c1*c2)*c3*v.*exp(1i*m*Phi);
Y{j} = sqrt(2)*real(Ylm);
elseif m>0
c1 = factorial(l-m)/factorial(l+m);
c2 = (2*l+1)/(4*pi);
v=reshape(p(abs(m)+1,:),[size(exp(1i*m*Phi),1),max(size(p(abs(m)+1,:),1),size(p(abs(m)+1,:),2))/size(exp(1i*m*Phi),1)])
Ylm = sqrt(c1*c2)*v.*exp(1i*m*Phi);
Y{j} = sqrt(2)*imag(Ylm);
else
c1 = factorial(l-m)/factorial(l+m);
c2 = (2*l+1)/(4*pi);
v=reshape(p(abs(m)+1,:),[size(exp(1i*m*Phi),1),max(size(p(abs(m)+1,:),1),size(p(abs(m)+1,:),2))/size(exp(1i*m*Phi),1)])
Ylm = sqrt(c1*c2)*v.*exp(1i*m*Phi);
Y{j} = Ylm;
end
end
end
all=zeros(61,121);
for q = 1:45
all = all+c(q)*Y{q};
end
[Xm,Ym,Zm] = sph2cart(Phi, pi/2-Theta,all);
surf(Xm,Ym,Zm)
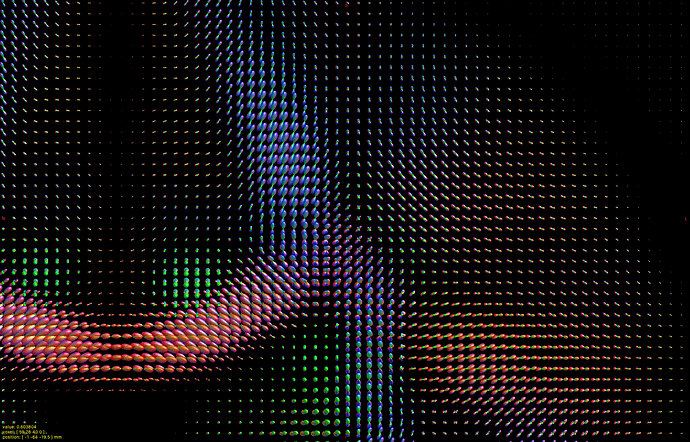
I hope this email finds you well. I am writing to seek your assistance regarding generating a visual FOD using 45 SH coefficients in Matlab.
I have already read the 45 SH coefficients in dimension 4 of the FOD, but I am unsure of how to use them to create a visual representation of the FOD in Matlab. As such, I am hoping that you could provide me with some guidance on how to go about this process.
Could you please advise me on the steps I should take to generate a visual FOD in Matlab using these 45 SH coefficients? It would be greatly appreciated if you could also provide me with any relevant code or resources that could help me with this task.
Thank you very much for your time and expertise. I look forward to hearing from you soon.
Best regards,
Ang