Hello,
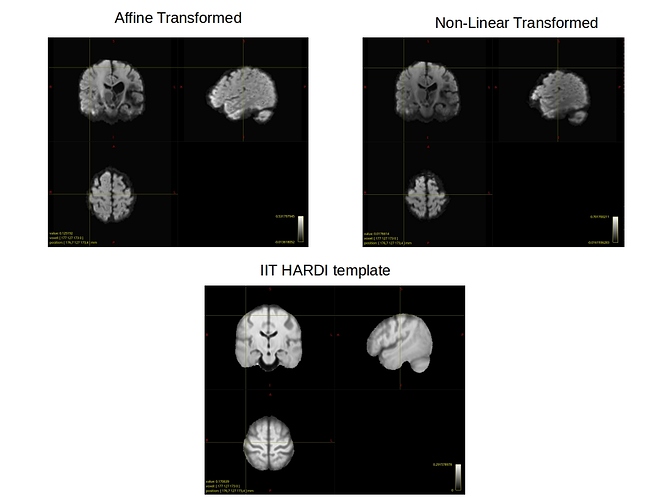
I am trying to use mrregister to register fod images of a patient population to the IIT HARDI template. I have encountered a problem that is rather strange. In particular, the registered images always end up just a bit smaller than the IIT HARDI template. The major subcortical structures are well aligned, but the cortex seems to have squeezed itself in. I believe this problem is coming from the non-linear part of the algorithm because when I run mrregister with just an affine transformation, the edges of the registered brain and the IIT HARDI template align quite well. It’s only when I run affine+nonlinear that my brains end up a bit smaller.
For my preprocessing, I have done denoising, topup and eddy, bias_correction, dwi2response dhollander, and dwi2fod msmt_csd. In particular, the image for which I am sending a screenshot was acquired with four shells (b=0,b=300,b=700,b=1500, > 100 directions total, and PA AP phase encoding). I am, however, observing the same behavior with data from a different subject, scanner, and acquisition parameters (b=0,b=1000, 30 direction). Since I would like to combine these populations, and the max lmax I can have is 4 (due to my population where only 30 directions are acquired), I have resampled the IIT HARDI template to lmax = 4 using sh2amp and amp2sh.
Here is my command for the non-linear transformation:
mrregister fod_wm.nii.gz IIT_HARDI_256_lmax4.nii.gz -force -transformed fod_wm_warped.nii.gz -nl_warp fod_wm_warp.nii.gz fod_wm_invwarp.nii.gz -mask1 fod_wm_mask.nii.gz -mask2 IITmean_tensor_mask_256.nii.gz
And here is my command for the only affine transformation
mrregister -force fod_wm.nii.gz IIT_HARDI_256_lmax4.nii.gz -type affine -transformed fod_wm_aff.nii.gz -mask1 fod_wm_mask.nii.gz -mask2 IITmean_tensor_mask_256.nii.gz
Here is a PDF where I show the curser on the IIT HARDI template, on my affine only transformed image, and on my nonlinear transformed image. As you can see, the nonlinear image is squeezed inward all around the brain, whereas the affine image aligns well with the size of the template. This can also be seen on the axial and sagittal planes where the brain appears smaller. I’m very confused as to what about the algorithm in the non-linear part is making my images shrink into the template, especially since the affine gives such a great initial alignment!
If anyone has used the IIT HARDI template for registering images and observed similar behavior or if anyone can give me any insight on this, I would be very appreciative.
Best regards,
Eric