Dear @jdtournier
There is a possible bug in the warpinit command (I don’t think it causes major problems … but it might)
In the message thread below, you state (see highlighted):
I’ve spent quite a bit of time making sure this issue with mismatched strides / transform in NIfTI images doesn’t happen - at least not routinely: NIfTI images produced by MRtrix3 should match the strides / transform of the input image, which should avoid these types of problems.
warpinit does not conform to this behaviour:
I ran this command:
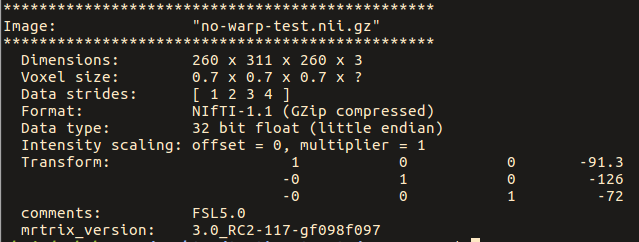
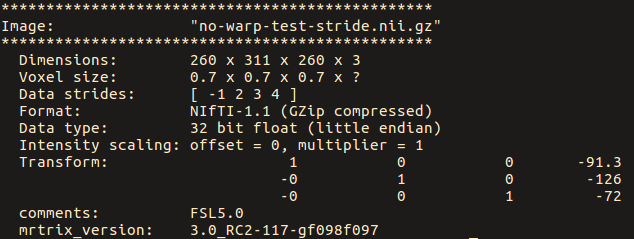
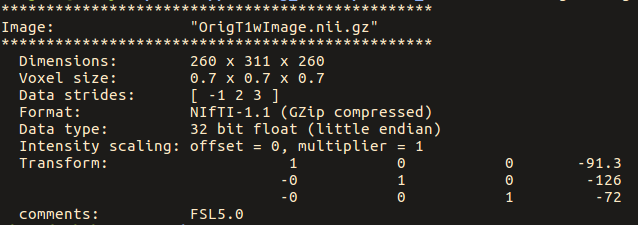
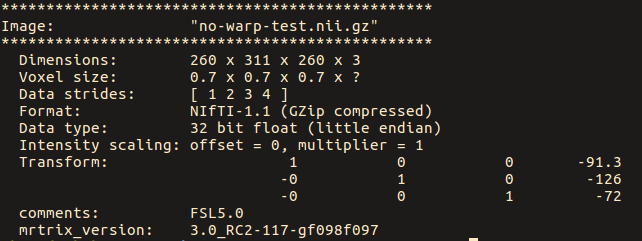
warpinit OrigT1wImage.nii.gz no-warp-test.nii.gz
Note below that the strides for the original image are: [ -1 2 3 ]
While for the ‘no-warp’ image, they are: [ 1 2 3 4 ]
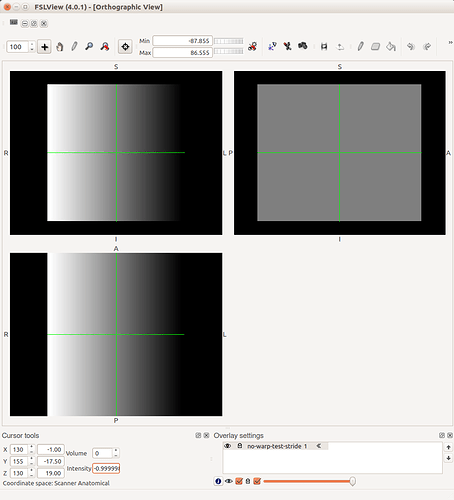
I have tested using this image for the to warp tracts to MNI space using fsl’s applywarp command:
warpinit OrigT1wImage.nii.gz no-warp-test.nii.gz
applywarp -i no-warp-test.nii.gz -r Native_T1w -w standard2acpc_dc.nii.gz -o warp.nii.gz
tcknormalise native_tract.tck warp.mif mni_tract.tck
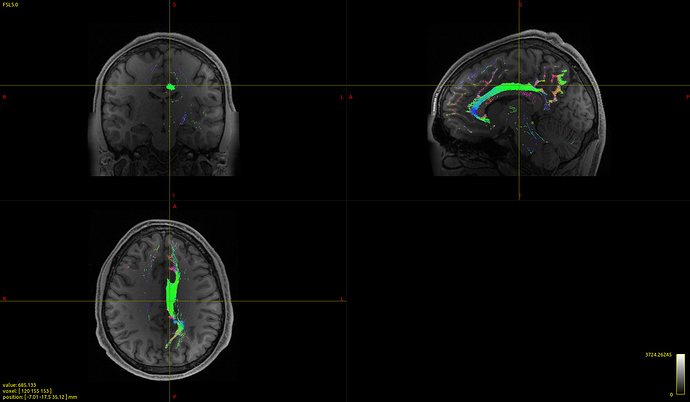
It seems to work with no problems and the registration of the tracts to the MNI brain look very good:
However, since you mention (in the above post) that:
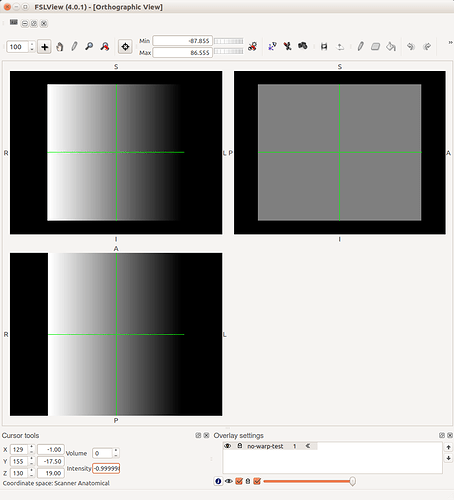
The only times you’ll need to worry about this is when interacting with other packages that may not handle these issues too well - or at least, handle them differently. One example is displaying an overlay on an image within FSLView
I thought I would bring up this issue.
Furthermore, I would like to also point to a thread on the FSL forum and a post by Mark Jenkinson that may answer some questions / explain why this was not actually causing a problem (though I suspect it will if someone simply loads the matrices into matlab or python and does not use real world coordinates):

source: JISCMail - FSL Archives
Interested to hear your thoughts on this matter.
Claude