Hi all,
I have some mutli-shell data (b0’s 700 (25dir), 1000 (40dir), and 2800 (75dir)), that I am processing with the help of mrtrix. After performing denoising, I calculate the residuals as suggested by the documentation.
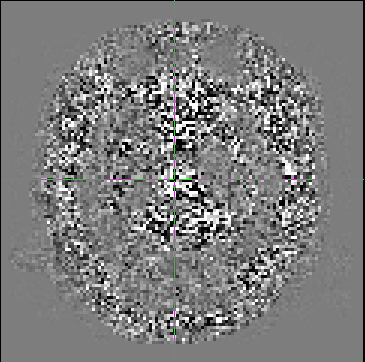
Although it doesn’t look as smooth as some examples I’ve seen previously (e.g http://community.mrtrix.org/t/dwidenoise-residuals-map/1293), there seems to be some anatomical information present.
I was hoping for some feedback as to whether this is correct/okay?
Cheers,
Forrest
Hi Forrest,
Apologies for the slow response – we’ve all been distracted by the ISMRM & subsequent MRtrix workshop…
I note your acquisition contains at least 25+40+75=135 (+ the b=0 volumes), which means the default spatial extent of 5×5×5=125 voxels will be smaller than the number of volumes. In this regime, it’s probably best to use (or at least try) a larger extent of 7×7×7. Try adding the -extent 7 to the dwidenoise command, see whether that helps.
Cheers,
Donald.
Hi Donald,
Thanks for the response; there’s no need to apologize!
I’ve tried using -extent 7 as suggested but am still getting similar residuals. If anything the features are more pronounced now.
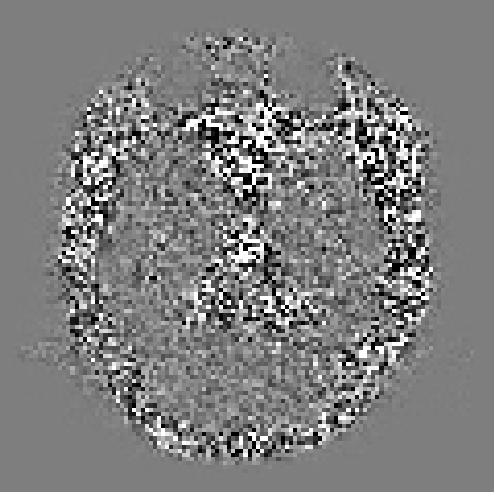
That is indeed pretty strange… The good news is that the residuals are smaller in brain parenchyma (and the eyes), so at least it’s not over-denoising. It’s not uncommon to see increased residuals in CSF, but I’m stumped as to why the residuals would be higher outside the brain…
A few things to check:
-
this is dwidenoise applied to the raw data straight off the scanner? Not following mrdegibbs or dwipreproc or anything like that? It really needs to be applied to the data before anything happens to it that might affect the noise distribution.
-
What does the noise map look like? Use the -noise option to get it.
-
The residuals will be a full 4D image, one per original volume in the data. Presumably we are looking at the first volume? What do the others look like? What does the RMS look like (you can compute that using mrmath residuals.mif rms -axis 3 rms_residuals.mif – edit as appropriate).
The residuals will be a full 4D image, one per original volume in the data. Presumably we are looking at the first volume? What do the others look like? What does the RMS look like (you can compute that using mrmath residuals.mif rms -axis 3 rms_residuals.mif – edit as appropriate).
I see now. You’re correct that was the first volume. I wasn’t aware that there were other volumes as well; although, with the name ‘residuals’ it seems pretty obvious in hindsight…
Thanks for you help and sorry to bother you with such a trivial problem!



