Hello everyone,
So here is the question…
I have diffusion data and I would like to do distortion correction… but I do not have two phase encoding directions, fieldmap or T2. Has anyone tried using a T1 for distortion correction? I have read a few papers about this topic but I was wondering if anyone has managed to implement this successfully?
Cheers,
Vasiliki
Hello
I did that a long time ago (for restropective data) and after testing different method, we found the most effective was to non-linearly register the csf segmented map (with spm you segment the T1 and then the mean B0) then you compute the deformation (with ants or other) between those csf map.
The advantage is that you have the same contrast.
At that time I did not find a way to constrain the non-linear deformation in the phase direction only. I think ants should be able to do it, (I do not know exactly how
Romain
Hi Vasiliki and Romain,
I had the same problem with my data, and what I did was rigid register the T1w to the B0, and after with ants non rigid register the B0 to the T1, restricting the registration with the -g option in ants. Finally apply the registration to all the volumes. It work for my data.
Regards,
Manuel
A common problem, and we had it as well with a lot of older, yet very important, historical data. I designed an algorithm specifically for this purpose just this year actually (and thanks to lots of work from @rtabbara, and lots of experimentation, we got it implemented as well). Our solution is based on some work I did during last year (see this abstract), where I was able to simulate the T1w image contrast quite closely from 3-tissue CSD results; but our iterative EPI distortion correction has some other bells and whistles now as well, like iteratively refined outlier rejection, and simultaneous (iterative) correction for both EPI distortions along the phase encoding direction on the end of the diffusion data combined with rigid motion correction of diffusion vs T1w data. The latter point is actually quite important: you’re after a combined model of EPI distortion as well as motion to be able to correct both accurately. They both depend on one another.
At the moment, this is just an internal prototype tuned for our own use cases (however, it did already run succesfully on a large cohort of 500+ subjects that often even have large lesions); but I’ll try and get something out at some point (the ISMRM deadline will be there surprisingly soon again any way; so that may just as well be a good opportunity). It’ll eventually make it into MRtrix3, but not any time (very) soon though, as I’ve got other priorities at the moment. If you think the other proposed solutions above wouldn’t cut it for your needs though, don’t hesitate to contact me privately: we can always arrange something if and when it makes sense.
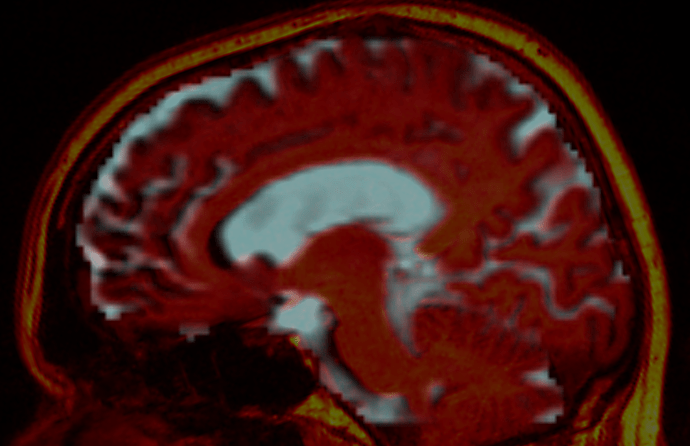
Here’s an example of a subject; before:
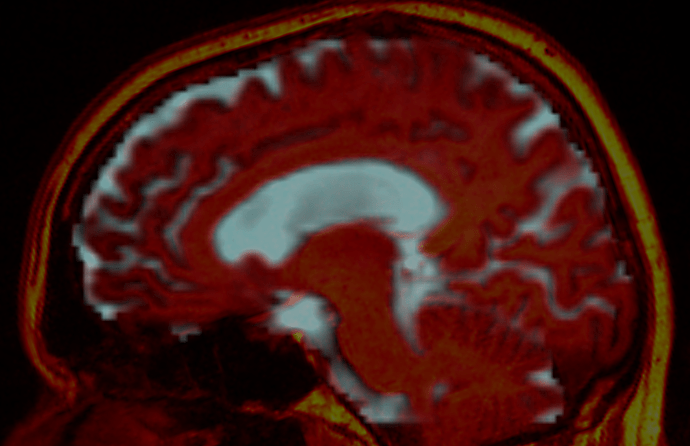
…and after:
For visualisation purposes, the CSF component from the 3-tissue CSD is shown to represent the diffusion data, overlaid by the T1w image in red (hot colour scale). The after image has the distortions corrected on the diffusion data, and the relative rigid motion corrected on the T1w image.
Quick question on this: doesn’t non-linearly deforming the DWI volumes affect the gradient directions (locally) ?
Good question. No, that shouldn’t affect the DW gradient directions, since these gradients are massive compared to the background gradients that cause the distortions. The phase-encoding gradient on the other hand is much smaller, at least when using EPI. The net result is the DW signal itself is not particularly affected, but it is misplaced.
Note that this is, maybe surprisingly at first sight, a reason that correction of these distortions is in a way extra important: these distortions don’t act like some sort of non-rigid motion on the images (i.e. as if the real brain was squished and stretched a bit in places), but just misplace the image data (signal) without adjusting the orientations accordingly. That means that, in the absence of correction for the distortions, the orientations (in some particular places) don’t line up any more with the surrounding spatial information; so a bundle can (in a way) be broken a bit. This may lead e.g. tractography to slightly struggle a bit more to make it through such bundles, since the local orientational information (of the FOD) may lead streamlines to bump into edges of structures, rather than traversing through.
Just to slightly add to that wording (even though meant completely correctly): note that the “misplaced” here means misplacement of absolute amounts of signal. What I mean to clarify by this, is that when the distortions non-linearly shrink a certain local region of the image, the signal gets added up (i.e. more signals get placed into a single voxel, if that makes sense). So regions that get squished, will appear brighter than they should’ve been; and regions that get stretched, will appear darker than they should’ve been. When applying a correction like the above, the image intensities should be modulated accordingly (based on the local determinant of the Jacobian of the warp).
Finally, with respect to those gradient directions again: if also correcting for rigid motion, that bit of the transformation, if applied to the diffusion data that is, should of course act on the gradient directions (since it’s genuine motion). However, since our approach over here (the one I illustrated above) acts on the FOD (and tissue) images directly, we would have to reorient the FODs. To avoid the computational cost and possible confusion related to that, I’ve designed that approach to correct the rigid motion “on the end of the T1”. It’s essentially all the same, since we’re only talking about relative motion between the 2 datasets (DWI and T1); but for some of our applications, it makes more sense to move the T1 rigidly around, rather than the DWI data. Since the T1w image is typically acquired first in most/all of our studies, we also see most motion happen “between the T1 and the rest of the acquisitions”, since subjects seem to move more (settle down more) initially. So correcting things like above, the T1 then often ends up being (magically) already much more aligned to the other acquisitions (e.g. a FLAIR or something) as well; which gives us more (and more robust) options to correct motion of those other acquisitions relative to all the rest.
Hi!
So, if somebody finds this useful:
Since SS3T-CSD is not currently implemented in MrTrix, I went ahead and wrote a small variant of it that seems to work quite well in simulating T1 contrast from DWI.
-
Following Wm ODF and response function with dhollander option _ single shell versus multi shell , I estimate response functions for CSF and WM only (I’m working on bad bad single-shell 30 gradient direction data)
-
I do linear registration between tissue probability maps max(P_csf, P_wm) (from the T1) and max(FA, ADC) (from DWI) (I forgot where but I think this was recommended somewhere on this forum as well=
-
Using that alignment, I follow Distortion correction using T1 to compute a linear mapping between [WM GM CSF] probabilities and T1 intensities - using the FA map instead of the GM response.
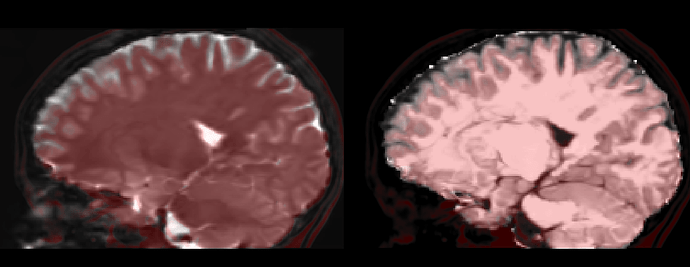
This seems to work quite well : left the b0, right the ‘simulated’ T1, both overlaid with the non-lineary registered T1 in red:
Dear all,
I was just wondering if there has been any progress in implementing non-linear registration to a T1 (or T1 and T2) in MRtrix or if there is a straightforward way to achieve this that you would recommend.
Thank you.
Is there any reason to use the warp field to modulate the DWI after it’s been normalized to the T1 image?
Hi
If you refer to the fact that one should modulate the DWI with the jacobian of the warp field I would say no :
The modulation is used when normalizing gray matter segmentation so that the deformation shrinking (or dilation) is reflected to the intensity of gray matter (used after for vbm).
but for diffusion the deformation is used to correct for the EPI distortion so we want to correct the deformation and keep the original intensity
Thanks. Just making sure my reasoning was correct on this.
Is there any reason to use the warp field to modulate the DWI after it’s been normalized to the T1 image?
Ideally, one would modulate the FODs based on any non-linear transfomation. Hopefully the figures in the original AFD manuscript should demonstrate why this is the case. However this modulation can’t be applied to the diffusion MRI intensities directly.
Any chance you have been able to repeat this process or document it in some way so that we could try and replicate it? I unfortunately have several old data sets with no opposite phase encoding directions that I have run through 3 tissue CSD.
Hey Daniel,
Yep, I’ve vaguely documented some ideas here a while ago: T1-like contrast from DWI data .
But I wouldn’t actually recommend to use this without a lot of experience in what’s going on here, as well as very careful QC. Generating the T1-like contrast itself is in some ways relatively straightforward actually, but it’s the registration for this particular purpose and sometimes in presence of certain particular pathologies, that is much harder to get right without a lot of experience in this area.
So while we did manage to get this right for a few particular studies at some point, I wouldn’t generally recommend to adopt this strategy broadly.
There’s a nice piece of work here though, and the software seems to be available: https://www.biorxiv.org/content/10.1101/2020.01.19.911784v1.full . It looks like quite an elegant approach with promising results. If you give it a go, I would actually love to hear if it works well for your data!
Cheers,
Thijs
There’s a nice piece of work here though, and the software seems to be available: https://www.biorxiv.org/content/10.1101/2020.01.19.911784v1.full . It looks like quite an elegant approach with promising results. If you give it a go, I would actually love to hear if it works well for your data!
Can confirm, Synb0-DISCO is fantastic and very comparable to topup.
Thanks for the feedback @lbinding. Is there any chance you have any documentation on running the docker scripts that are available for Synb0-DISCO.
I have docker installed however I find the documentation on running a bit confusing.
Specifically, if I have an input folder with the requested documents.
How do I set up this command?
sudo docker run --rm \
-v $(pwd)/INPUTS/:/INPUTS/ \
-v $(pwd)/OUTPUTS:/OUTPUTS/ \
-v <path to license.txt>:/extra/freesurfer/license.txt \
--user $(id -u):$(id -g) \
justinblaber/synb0_25iso
I have
@CallowBrainProject No worries, I’ve been thinking about writing up my pipeline on here as I’ve spent the last 2 months trying to get optimal tractography for older and newer data… I’m sure I’ll get round to it at some point…
So I have a folder in each participant called synb0 where I have my T1, the b0 and acqparams. The acqparams need to be changed so it has another line i.e. if the original acqparams.txt file is:
0 1 0 0.035225
you need to add another line with all zeros on the 4th entry i.e:
0 1 0 0.035225
0 1 0 0.000000
After that I would call synb0-disco like:
sudo docker run --rm
-v /Users/lbinding/Desktop/subjects/161/synb0/:/INPUTS/
-v /Users/lbinding/Desktop/subjects/161/synb0/OUTPUTS:/OUTPUTS/
-v /Applications/freesurfer/license.txt:/extra/freesurfer/license.txt
–user $(id -u):$(id -g)
justinblaber/synb0_25iso
Hope that helps, if you have issues I’m more than happy to help where I can.
Hi all - I’m glad this tool has been useful for several groups!
@CallowBrainProject, just to clarify here, when you run this command you are running the docker image (named synb0_25iso) that is essentially a system-agnostic black box that is only good (in our case) for running this one specific process. You can think of this as each line in the above command is binding a directory on your system to a directory within this black-box. So anything before the “:” should point to the inputs on your file system and everything after the “:” remains unchanged. For example we choose to literally create a folder named INPUTS but you can use any folder on your system as an inputs folder and bind it to the /INPUTS/ directory within the docker image.
The issue that we’ve run into most is the that should point to a free surfer license.txt file (you get this when you download free surfer). For example, my license file sits in /Applications/freesurfer/ so my line would look like:
-v /Applications/freesurfer/:/extra/freesurfer/license.txt
We use a freesurfer command during a preprocessing step and are looking to eliminate this in a future iteration to make this more simple for users.
Also - this docker/singularity image will create a synthesized image and run TOPUP for you. You will then want to use the output (movement parameters and field coefficients) with EDDY in exactly the same way you would if you had started with a full set of blip-up blip-down acquisitions. We are trying to update the GitHub to make this very intuitive to use (I apologize this is my first tool I’ve released and I can only imagine how hard it would be to maintain an entire software framework like MRtrix3!).
One last note, we would recommend after running this pipeline to do a visual inspection of the b0_all.nii.gz and flip back-and-forth between the first and second volumes. The first one is your distortion-corrected b0 which should now match the second volume (the synthesized b0 that is undistorted) exactly!
Would love any feedback on the output or running the image itself!
Hello, thank you so much for all the input and advice. I am really excited to try and run this.
I am currently running the following!
sudo docker run --rm \
-v /Volumes/DANIEL/EPC/analysis/EPC001.post/input:/INPUTS/ \
-v /Volumes/DANIEL/EPC/analysis/EPC001.post/output:/OUTPUTS/ \
-v /Applications/freesurfer_dev/license.txt:/extra/freesurfer/license.txt \
-user $(id -u):$(id -g) \
justinblaber/synbo_25iso
However, I get the following error when trying to run…
Unable to find image ‘501:20’ locally
docker: Error response from daemon: pull access denied for 501, repository does not exist or may require ‘docker login’: denied: requested access to the resource is denied.
See ‘docker run --help’.