Hi, J-Donald,
I have done several tests on the data with your suggestion.
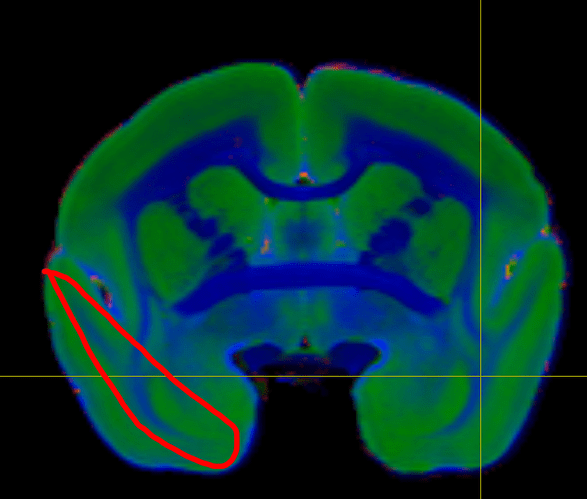
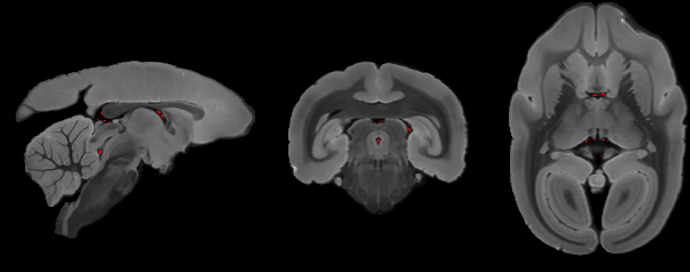
It did a surprisingly good job in segmenting the gray matter and white matter of the marmoset brain. See the thin white matter in the temporal-pole area. Previously, I have tested traditional segmentation tools (fast, ants) Neither of them fully segment-out this thin white matter using MTR (T1) and T2 ex-vivo image. But, the FOD of mrtrix3 did!
--------------What I did
_ To generate response functions, here was what I did to select voxels:_
a. White matter - I used two methods. One was as you suggested, I manually chose voxels with highest FA (top 300). Second is using single shell (7200) and applying Tournier method to obtain the white matter voxels (300), and then used the voxels for manual multi-shell dwi2response.
(the blue is manual and the red is Tournier)
b. Gray matter - The marmoset cortex is too thick and contain many voxels. Thus, I manually selected about 1500 voxels with FA lower than 0.1. Note that with the high-resolution ex-vivo data and relative thick cortex of marmoset, we can see the different layers of the cortex that have different FA value and features. This is different from what we have on human invivo data. I am not sure whether this simple method is appropriate or not.
c. CSF - The ventricle of marmoset is not that big. So I manually selected 150 voxels of the CSF from S0 image.
Then I did what you suggested (lmax = 0,4,6,8 for the white matter of different shells; lmax = 0,0,0,0 for gm&csf ). Besides the encouraging results, there are still issues that I am not quite confident:
--------------Issues
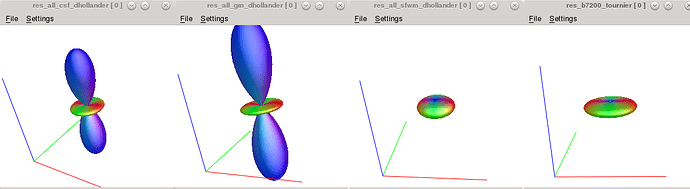
1. Issue of b=7200 again:
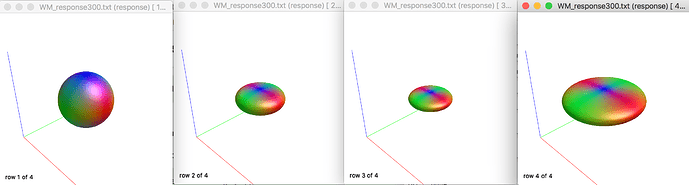
(from left to right: b0 (lmax0), b2400 (lmax4), b4800 (lmax 6), b7200 (lmax8))
Why the radius of circles of b4800 is smaller than b2400, but b7200 is larger than all lower b value? (the response value is listed in the next question)
Is this normal?
As I mentioned previously, once b=7200 involved, the FA of many white matter will >1. I have used the right Receiver Gain to adjust the data, still the same results.
I then consulted expertises in DTI, and identified the reason:
"(Niethammer, et al, 2006, On Diffusion Tensor Estimation) Simple tensor fitting could give rise negative eigenvalues: Because dMRI signal can be really small at b=7200 when G is parallel to a fiber, the fitting can be very sensitive to noise when noise level is in fact above the minimum signal level. The formulation of FA makes it larger than 1 when some EV is negative. ".
Now, for the Mrtrix and the CSD tracking, etc, will it be affected by the issue as the tensor model?
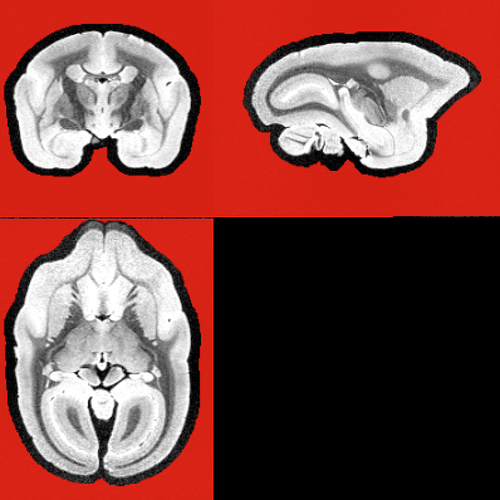
2. Manually selected vs. tournier:
The Tournier selected many different voxels than the FA-manual method and resulting different response functions (for 300 white matter voxels).
(left: manual; right:tournier)
The touriner has smaller (radius) but flatter shape than the manual.
Which one is better?
**_Manual:_**
_173670.8311645914 0 0 0 0_
_112471.0979004641 -30904.53437063165 6914.149390457013 0 0_
_84556.50252458362 -30573.53020595047 12082.37401380716 -3937.906991237937 0_
_146785.459380438 -59106.35423326885 26799.40599680597 -11027.75365967198 2882.521653928761_
**_Touriner:_**
_322084.1663155202 0 0 0 0_
_183785.3993442675 -59540.47745553248 13320.75581448077 0 0_
_122420.6147702928 -53843.52800533723 23477.13836021282 -8166.972327980991 0_
_212011.0697191145 -100392.5019609229 50932.57994245004 -23922.30182768965 6450.961159453199_
3. About FOD of gray matter and CSF
For tckgen, only the white matter FOD are needed.
What is the usage of FOD of gm and csf?
4. Track Orientation Density Imaging (TODI)
I have read the paper about the TODI and also watched the concept video on youtube about the TODI.
The super-resolution image generated will be extremely useful for animal data with thin white matter, including rodents and marmosets. I want follow the concept in the TODI paper and try to implement it on the marmoset data. However, the discussion about implementation I in the paper is general.
Are there any guidelines and suggestions for implementing the new method?
There is a command, tckmap, able to generate old TDI and the new TODI (a lmax can be defined).
Should I use higher lmax for dwi2response and dwi2fod (in the paper, lmax upto 16 was tested for TODI).
Thanks in advance!