mblesac
October 18, 2020, 12:14pm
1
Dear Mrtrix experts,
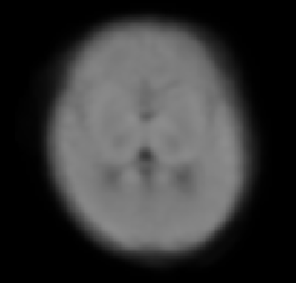
I’m running population_template on my neonatal population, the first average looks reasonable:
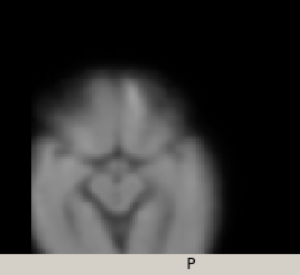
however the template is outside the FOV, I found that the error is in the first rigid registration:
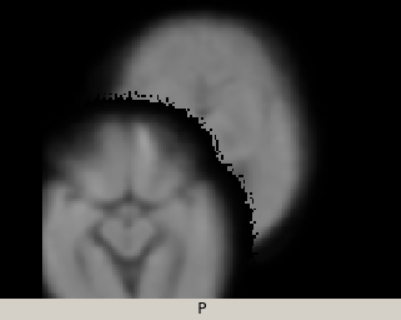
And both overlaid:
Is there any way to optimize this first step in the population_template script? Thanks in advance.
Best regards,
Manuel
1 Like
Hi Manuel,
This looks like an issue with the initial alignment (-inital_alignment) before any linear registration is run but given your description I am having trouble seeing what exactly the issue might be. One option is that none of your masks cover the entire brain (check with mrview image -roi.load mask). Otherwise, to debug this, could you check which files in the scratch directory are fine and what the first cropped image is in initial_template*.mif linear_template*.mif?
Hi Max,
Thanks for your reply. I tried changing the -initial_alignment to robust_mass and the result is the same. I inspected all the masks and they look ok.
These images were converted from dicom to nifti long ago. See attached the outputs from mrinfo and fslhd for one subject:
Here the output of mrinfo:
************************************************
Image name: "7005/7005-dwi_den_eddy_bias_res.nii.gz"
************************************************
Dimensions: 256 x 256 x 100 x 75
Voxel size: 1 x 1 x 1 x 1
Data strides: [ -1 2 3 4 ]
Format: NIfTI-1.1 (GZip compressed)
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 -254.5
-0 1 0 -0.5
-0 0 1 -0.5
comments: 5.0.11
mrtrix_version: 3.0_RC3-15-g9494da8d
and the output from fslhd:
filename 7005/7005-dwi_den_eddy_bias_res.nii.gz
sizeof_hdr 348
data_type FLOAT32
dim0 4
dim1 256
dim2 256
dim3 100
dim4 75
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim0 -1.000000
pixdim1 1.000000
pixdim2 1.000000
pixdim3 1.000000
pixdim4 1.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -1.000000 0.000000 -0.000000 0.500000
qto_xyz:2 0.000000 1.000000 -0.000000 -0.500000
qto_xyz:3 0.000000 0.000000 1.000000 -0.500000
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -1.000000 0.000000 0.000000 0.500000
sto_xyz:2 0.000000 1.000000 0.000000 -0.500000
sto_xyz:3 0.000000 0.000000 1.000000 -0.500000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip MRtrix version: 3.0_RC3-15-g9494da8d
aux_file
I find strange the last column of the qform to be 0 0 0 1 (in the not resampled image) but I don’t know if this is related at all with the issue.
Best regards,
Manuel
Just to rule out a viewer problem, have you looked at the images with mrview? Which of the following images is cropped?
population_template <odfs> out.mif -mask <masks> -type rigid -rigid_scale 0.3,0.4 -scratch regtest -nocleanup
cd regtest
mrview -load initial_template_c0.mif -colourmap 4 -load linear_template00_c0.mif -colourmap 4 -load linear_template01_c0.mif -colourmap 4 -mode 2
I don’t think I can figure out what’s going on with the header information only. If you can reproduce the problem with a few images and are able to share these and the command options that’d probably the fastest way to get to the bottom of this.
Otherwise, I’d modify population_template and replace this line by
average_inv = None
and check if the problem goes away. If so I’d be interested in the content of the linear_transforms_* folders in the scratch dir generated with this modification.
1 Like
Hi Max,
I can see the same problem in two different versions of the software, the 3.0_RC3-15-g9494da8d and the 3.0.0
The FOD images look right in mrview
When I run this:
population_template out.mif -mask -type rigid -rigid_scale 0.3,0.4 -scratch regtest -nocleanup
I find the following error:
population_template: Generating a population-average template from 20 input images
population_template: SH Series detected, performing FOD registration in contrast: regtest/fod_input
population_template: [ERROR] Unhandled Python exception:
population_template: [ERROR] AssertionError:
population_template: [ERROR] Traceback:
population_template: [ERROR] /EBC/local/MRtrix3_stable/mrtrix3/bin/population_template:823 (in execute())
population_template: [ERROR] assert len(linear_lmax) == len(linear_niter)
I’ll try to find a way to share the images.
Best regards,
Manuel
mblesac
October 22, 2020, 11:35am
6
Hi Max,
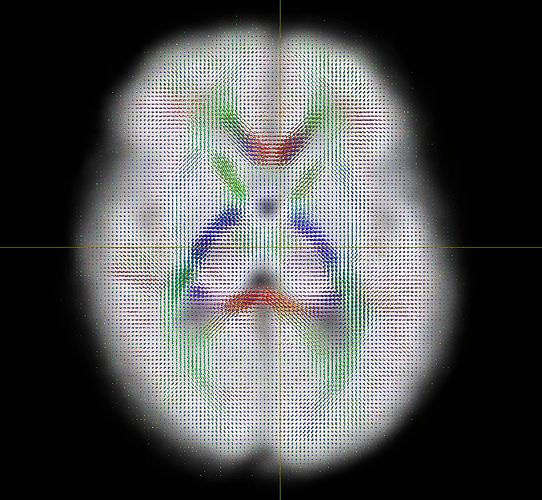
After modifing the line as you suggested and running the following command:
population_template <odfs> out.mif -mask <masks> -type rigid -scratch regtest -nocleanup
I obtained the following template
Do you want to take a look to the linear_transforms_* folders? You can see the files here where the modified are the ones with the modified version of population_template (the password is template). Thanks in advance.
Best regards,
Manuel
1 Like