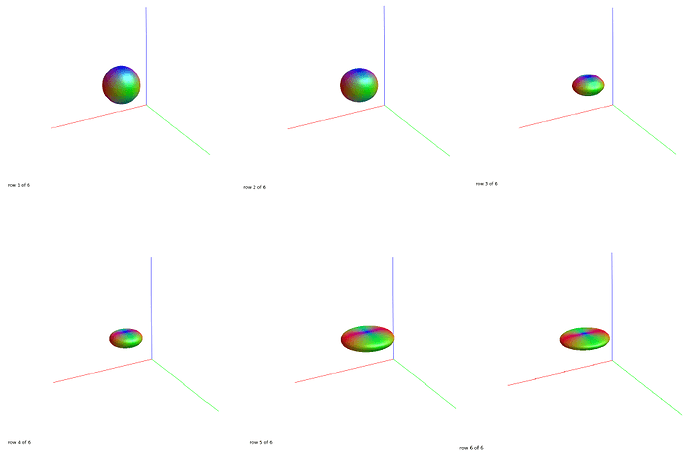
Hi @jdtournier,
thank you for the quick response. As requested, you can find the line command and all of the outputs below :
dwi2response msmt_5tt -voxels voxels.mif -info ../01-MifConversion/dwi.mif ../01-MifConversion/5TT/5TT.mif WM_response.txt GM_response.txt CSF_response.txt
dwi2response:
dwi2response: Note that this script makes use of commands / algorithms that have relevant articles for citation. Please consult the help page (-help option) for more information.
dwi2response:
dwi2response: Command: '/home/ateillac/Softs/MRTRIX/mrtrix3/bin/mrinfo /scratch/ateillac/NHP/M157_5shells/Essai/01-MifConversion/dwi.mif -dwgrad' (piping data to local storage)
dwi2response: Result: (218 lines)
dwi2response: Generated temporary directory: /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/
Command: mrconvert /scratch/ateillac/NHP/M157_5shells/Essai/01-MifConversion/dwi.mif /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi.mif -stride 0,0,0,1
mrconvert: [100%] copying from "/scratch/a...ai/01-MifConversion/dwi.mif" to "/scratch/a...response-tmp-DRG9XQ/dwi.mif"
Command: mrconvert /scratch/ateillac/NHP/M157_5shells/Essai/01-MifConversion/5TT/5TT.mif /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/5tt.mif
mrconvert: [100%] copying from "/scratch/a...1-MifConversion/5TT/5TT.mif" to "/scratch/a...response-tmp-DRG9XQ/5tt.mif"
dwi2response: Changing to temporary directory (/scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/)
Command: dwi2mask dwi.mif mask.mif
dwi2mask: [100%] finding min/max of "mean b=0 image"
dwi2mask: [done] optimising threshold
dwi2mask: [100%] thresholding
dwi2mask: [100%] finding min/max of "mean b=244 image"
dwi2mask: [done] optimising threshold
dwi2mask: [100%] thresholding
dwi2mask: [100%] finding min/max of "mean b=800 image"
dwi2mask: [done] optimising threshold
dwi2mask: [100%] thresholding
dwi2mask: [100%] finding min/max of "mean b=1536 image"
dwi2mask: [done] optimising threshold
dwi2mask: [100%] thresholding
dwi2mask: [100%] finding min/max of "mean b=4075 image"
dwi2mask: [done] optimising threshold
dwi2mask: [100%] thresholding
dwi2mask: [100%] finding min/max of "mean b=7106 image"
dwi2mask: [done] optimising threshold
dwi2mask: [100%] thresholding
dwi2mask: [done] computing dwi brain mask
dwi2mask: [done] applying mask cleaning filter
Command: 5ttcheck 5tt.mif
dwi2response: Command: '/home/ateillac/Softs/MRTRIX/mrtrix3/bin/mrinfo dwi.mif -shells' (piping data to local storage)
dwi2response: Result: 0 243.7 800.471 1536.39 4075.06 7106.2
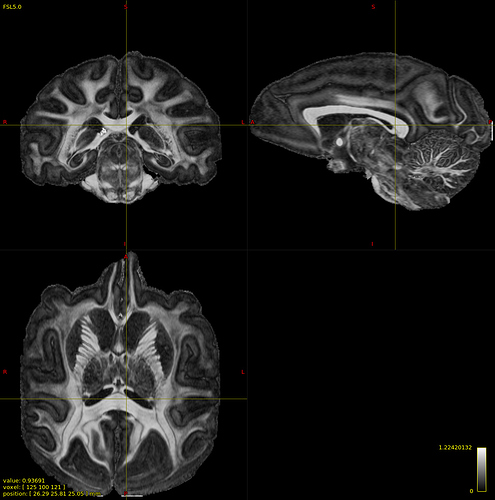
Command: dwi2tensor dwi.mif - -mask mask.mif | tensor2metric - -fa fa.mif -vector vector.mif
dwi2tensor: [100%] computing tensors
tensor2metric: [100%] computing metrics
Function: shutil.copy(vector.mif, dirs.mif)
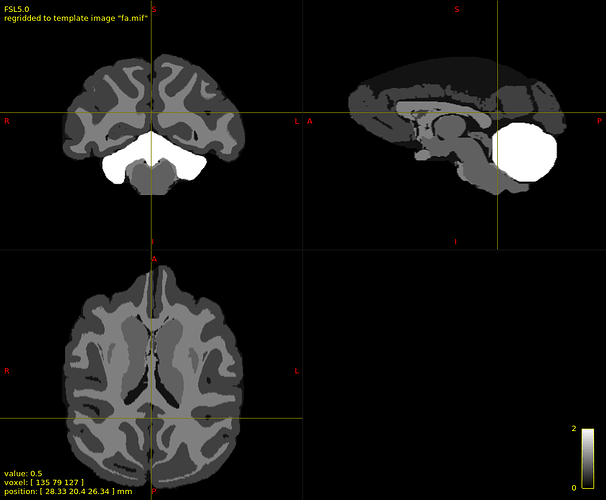
Command: mrtransform 5tt.mif 5tt_regrid.mif -template fa.mif -interp linear
mrtransform: [100%] reslicing "5tt.mif"
Command: mrconvert 5tt_regrid.mif - -coord 3 2 -axes 0,1,2 | mrcalc - 0.95 -gt mask.mif -mult wm_mask.mif
mrconvert: [100%] copying from "5tt_regrid.mif" to "/tmp/mrtrix-tmp-2Kvuz0.mif"
mrcalc: [100%] computing: ((/tmp/mrtrix-tmp-2Kvuz0.mif > 0.95) * mask.mif)
Command: mrconvert 5tt_regrid.mif - -coord 3 0 -axes 0,1,2 | mrcalc - 0.95 -gt fa.mif 0.2 -lt -mult mask.mif -mult gm_mask.mif
mrconvert: [100%] copying from "5tt_regrid.mif" to "/tmp/mrtrix-tmp-2Kvuz0.mif"
mrcalc: [100%] computing: (((/tmp/mrtrix-tmp-2Kvuz0.mif > 0.95) * (fa.mif < 0.2)) * mask.mif)
Command: mrconvert 5tt_regrid.mif - -coord 3 3 -axes 0,1,2 | mrcalc - 0.95 -gt fa.mif 0.2 -lt -mult mask.mif -mult csf_mask.mif
mrconvert: [100%] copying from "5tt_regrid.mif" to "/tmp/mrtrix-tmp-2Kvuz0.mif"
mrcalc: [100%] computing: (((/tmp/mrtrix-tmp-2Kvuz0.mif > 0.95) * (fa.mif < 0.2)) * mask.mif)
dwi2response: Calling dwi2response recursively to select WM single-fibre voxels using 'tournier' algorithm
Command: dwi2response tournier dwi.mif wm_ss_response.txt -mask wm_mask.mif -voxels wm_sf_mask.mif -tempdir /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/
dwi2response:
dwi2response: Note that this script makes use of commands / algorithms that have relevant articles for citation. Please consult the help page (-help option) for more information.
dwi2response:
dwi2response: Generated temporary directory: /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi2response-tmp-BWAOF1/
Command: mrconvert /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi.mif - -stride 0,0,0,1 | dwiextract - /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi2response-tmp-BWAOF1/dwi.mif -singleshell -no_bzero
Command: mrconvert /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/wm_mask.mif /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi2response-tmp-BWAOF1/mask.mif -datatype bit
dwi2response: Changing to temporary directory (/scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi2response-tmp-BWAOF1/)
Command: dwi2fod csd dwi.mif init_RF.txt iter0_FOD.mif -mask mask.mif -lmax 4
Command: fod2fixel iter0_FOD.mif iter0_fixel -peak peaks.mif -mask mask.mif -fmls_no_thresholds
Command: fixel2voxel iter0_fixel/peaks.mif split_data iter0_amps.mif -number 2
Command: mrconvert iter0_amps.mif iter0_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter0_amps.mif iter0_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter0_fixel/directions.mif split_dir iter0_all_dirs.mif -number 1
Command: mrconvert iter0_all_dirs.mif iter0_first_dir.mif -coord 3 0:2
Command: mrcalc iter0_first_peaks.mif -sqrt 1 iter0_second_peaks.mif iter0_first_peaks.mif -div -sub 2 -pow -mult iter0_CF.mif
Command: mrthreshold iter0_CF.mif -top 300 iter0_SF.mif
Command: amp2response dwi.mif iter0_SF.mif iter0_first_dir.mif iter0_RF.txt -lmax 4
Command: mrthreshold iter0_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter0_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter0_RF.txt iter1_FOD.mif -mask iter0_SF_dilated.mif
Command: fod2fixel iter1_FOD.mif iter1_fixel -peak peaks.mif -mask iter0_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter1_fixel/peaks.mif split_data iter1_amps.mif -number 2
Command: mrconvert iter1_amps.mif iter1_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter1_amps.mif iter1_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter1_fixel/directions.mif split_dir iter1_all_dirs.mif -number 1
Command: mrconvert iter1_all_dirs.mif iter1_first_dir.mif -coord 3 0:2
Command: mrcalc iter1_first_peaks.mif -sqrt 1 iter1_second_peaks.mif iter1_first_peaks.mif -div -sub 2 -pow -mult iter1_CF.mif
Command: mrthreshold iter1_CF.mif -top 300 iter1_SF.mif
Command: amp2response dwi.mif iter1_SF.mif iter1_first_dir.mif iter1_RF.txt
Command: mrcalc iter1_SF.mif iter0_SF.mif -sub iter1_SF_diff.mif
Command: mrthreshold iter1_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter1_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter1_RF.txt iter2_FOD.mif -mask iter1_SF_dilated.mif
Command: fod2fixel iter2_FOD.mif iter2_fixel -peak peaks.mif -mask iter1_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter2_fixel/peaks.mif split_data iter2_amps.mif -number 2
Command: mrconvert iter2_amps.mif iter2_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter2_amps.mif iter2_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter2_fixel/directions.mif split_dir iter2_all_dirs.mif -number 1
Command: mrconvert iter2_all_dirs.mif iter2_first_dir.mif -coord 3 0:2
Command: mrcalc iter2_first_peaks.mif -sqrt 1 iter2_second_peaks.mif iter2_first_peaks.mif -div -sub 2 -pow -mult iter2_CF.mif
Command: mrthreshold iter2_CF.mif -top 300 iter2_SF.mif
Command: amp2response dwi.mif iter2_SF.mif iter2_first_dir.mif iter2_RF.txt
Command: mrcalc iter2_SF.mif iter1_SF.mif -sub iter2_SF_diff.mif
Command: mrthreshold iter2_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter2_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter2_RF.txt iter3_FOD.mif -mask iter2_SF_dilated.mif
Command: fod2fixel iter3_FOD.mif iter3_fixel -peak peaks.mif -mask iter2_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter3_fixel/peaks.mif split_data iter3_amps.mif -number 2
Command: mrconvert iter3_amps.mif iter3_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter3_amps.mif iter3_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter3_fixel/directions.mif split_dir iter3_all_dirs.mif -number 1
Command: mrconvert iter3_all_dirs.mif iter3_first_dir.mif -coord 3 0:2
Command: mrcalc iter3_first_peaks.mif -sqrt 1 iter3_second_peaks.mif iter3_first_peaks.mif -div -sub 2 -pow -mult iter3_CF.mif
Command: mrthreshold iter3_CF.mif -top 300 iter3_SF.mif
Command: amp2response dwi.mif iter3_SF.mif iter3_first_dir.mif iter3_RF.txt
Command: mrcalc iter3_SF.mif iter2_SF.mif -sub iter3_SF_diff.mif
Command: mrthreshold iter3_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter3_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter3_RF.txt iter4_FOD.mif -mask iter3_SF_dilated.mif
Command: fod2fixel iter4_FOD.mif iter4_fixel -peak peaks.mif -mask iter3_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter4_fixel/peaks.mif split_data iter4_amps.mif -number 2
Command: mrconvert iter4_amps.mif iter4_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter4_amps.mif iter4_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter4_fixel/directions.mif split_dir iter4_all_dirs.mif -number 1
Command: mrconvert iter4_all_dirs.mif iter4_first_dir.mif -coord 3 0:2
Command: mrcalc iter4_first_peaks.mif -sqrt 1 iter4_second_peaks.mif iter4_first_peaks.mif -div -sub 2 -pow -mult iter4_CF.mif
Command: mrthreshold iter4_CF.mif -top 300 iter4_SF.mif
Command: amp2response dwi.mif iter4_SF.mif iter4_first_dir.mif iter4_RF.txt
Command: mrcalc iter4_SF.mif iter3_SF.mif -sub iter4_SF_diff.mif
Command: mrthreshold iter4_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter4_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter4_RF.txt iter5_FOD.mif -mask iter4_SF_dilated.mif
Command: fod2fixel iter5_FOD.mif iter5_fixel -peak peaks.mif -mask iter4_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter5_fixel/peaks.mif split_data iter5_amps.mif -number 2
Command: mrconvert iter5_amps.mif iter5_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter5_amps.mif iter5_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter5_fixel/directions.mif split_dir iter5_all_dirs.mif -number 1
Command: mrconvert iter5_all_dirs.mif iter5_first_dir.mif -coord 3 0:2
Command: mrcalc iter5_first_peaks.mif -sqrt 1 iter5_second_peaks.mif iter5_first_peaks.mif -div -sub 2 -pow -mult iter5_CF.mif
Command: mrthreshold iter5_CF.mif -top 300 iter5_SF.mif
Command: amp2response dwi.mif iter5_SF.mif iter5_first_dir.mif iter5_RF.txt
Command: mrcalc iter5_SF.mif iter4_SF.mif -sub iter5_SF_diff.mif
Command: mrthreshold iter5_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter5_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter5_RF.txt iter6_FOD.mif -mask iter5_SF_dilated.mif
Command: fod2fixel iter6_FOD.mif iter6_fixel -peak peaks.mif -mask iter5_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter6_fixel/peaks.mif split_data iter6_amps.mif -number 2
Command: mrconvert iter6_amps.mif iter6_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter6_amps.mif iter6_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter6_fixel/directions.mif split_dir iter6_all_dirs.mif -number 1
Command: mrconvert iter6_all_dirs.mif iter6_first_dir.mif -coord 3 0:2
Command: mrcalc iter6_first_peaks.mif -sqrt 1 iter6_second_peaks.mif iter6_first_peaks.mif -div -sub 2 -pow -mult iter6_CF.mif
Command: mrthreshold iter6_CF.mif -top 300 iter6_SF.mif
Command: amp2response dwi.mif iter6_SF.mif iter6_first_dir.mif iter6_RF.txt
Command: mrcalc iter6_SF.mif iter5_SF.mif -sub iter6_SF_diff.mif
Command: mrthreshold iter6_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter6_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter6_RF.txt iter7_FOD.mif -mask iter6_SF_dilated.mif
Command: fod2fixel iter7_FOD.mif iter7_fixel -peak peaks.mif -mask iter6_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter7_fixel/peaks.mif split_data iter7_amps.mif -number 2
Command: mrconvert iter7_amps.mif iter7_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter7_amps.mif iter7_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter7_fixel/directions.mif split_dir iter7_all_dirs.mif -number 1
Command: mrconvert iter7_all_dirs.mif iter7_first_dir.mif -coord 3 0:2
Command: mrcalc iter7_first_peaks.mif -sqrt 1 iter7_second_peaks.mif iter7_first_peaks.mif -div -sub 2 -pow -mult iter7_CF.mif
Command: mrthreshold iter7_CF.mif -top 300 iter7_SF.mif
Command: amp2response dwi.mif iter7_SF.mif iter7_first_dir.mif iter7_RF.txt
Command: mrcalc iter7_SF.mif iter6_SF.mif -sub iter7_SF_diff.mif
Command: mrthreshold iter7_CF.mif -top 3000 - | maskfilter - dilate - -npass 1 | mrcalc mask.mif - -mult iter7_SF_dilated.mif
Command: dwi2fod csd dwi.mif iter7_RF.txt iter8_FOD.mif -mask iter7_SF_dilated.mif
Command: fod2fixel iter8_FOD.mif iter8_fixel -peak peaks.mif -mask iter7_SF_dilated.mif -fmls_no_thresholds
Command: fixel2voxel iter8_fixel/peaks.mif split_data iter8_amps.mif -number 2
Command: mrconvert iter8_amps.mif iter8_first_peaks.mif -coord 3 0 -axes 0,1,2
Command: mrconvert iter8_amps.mif iter8_second_peaks.mif -coord 3 1 -axes 0,1,2
Command: fixel2voxel iter8_fixel/directions.mif split_dir iter8_all_dirs.mif -number 1
Command: mrconvert iter8_all_dirs.mif iter8_first_dir.mif -coord 3 0:2
Command: mrcalc iter8_first_peaks.mif -sqrt 1 iter8_second_peaks.mif iter8_first_peaks.mif -div -sub 2 -pow -mult iter8_CF.mif
Command: mrthreshold iter8_CF.mif -top 300 iter8_SF.mif
Command: amp2response dwi.mif iter8_SF.mif iter8_first_dir.mif iter8_RF.txt
Command: mrcalc iter8_SF.mif iter7_SF.mif -sub iter8_SF_diff.mif
dwi2response: Convergence of SF voxel selection detected at iteration 8
Function: shutil.copyfile(iter8_RF.txt, response.txt)
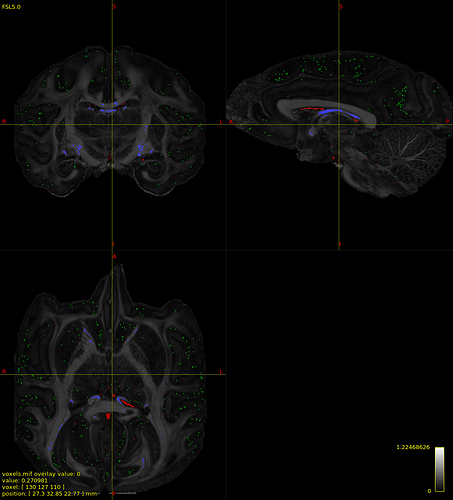
Function: shutil.move(iter8_SF.mif, voxels.mif)
Function: shutil.copyfile(response.txt, /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/wm_ss_response.txt)
Command: mrconvert voxels.mif /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/wm_sf_mask.mif
dwi2response: Changing back to original directory (/scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ)
dwi2response: Deleting temporary directory /scratch/ateillac/NHP/M157_5shells/Essai/02-TissueResponse/dwi2response-tmp-DRG9XQ/dwi2response-tmp-BWAOF1/
dwi2response: Command: '/home/ateillac/Softs/MRTRIX/mrtrix3/bin/mrstats wm_sf_mask.mif -output count -mask wm_sf_mask.mif' (piping data to local storage)
dwi2response: Result: 300
dwi2response: Command: '/home/ateillac/Softs/MRTRIX/mrtrix3/bin/mrstats gm_mask.mif -output count -mask gm_mask.mif' (piping data to local storage)
dwi2response: Result: 1564410
dwi2response: Command: '/home/ateillac/Softs/MRTRIX/mrtrix3/bin/mrstats csf_mask.mif -output count -mask csf_mask.mif' (piping data to local storage)
dwi2response: Result: 134401
Command: amp2response dwi.mif wm_sf_mask.mif dirs.mif wm.txt -shell 0,244,800,1536,4075,7106
amp2response: Shell b=0: Response function [ 20490.2344 ] from average of 300 voxels
amp2response: Shell b=244: Response function [ 19293.3223 -1157.88391 -629.721313 -75.8945007 108.068123 48.0639648 ] solved after 9 constraint iterations from 300 voxels
amp2response: Shell b=800: Response function [ 14563.3799 -2763.39209 46.4419518 -19.7071095 -32.4169006 -50.580658 ] solved after 0 constraint iterations from 300 voxels
amp2response: Shell b=1536: Response function [ 13083.3184 -3678.49072 743.963013 -121.122055 35.3785896 1.31172085 ] solved after 0 constraint iterations from 300 voxels
amp2response: Shell b=4075: Response function [ 8740.21289 -3971.81079 1678.5708 -571.056824 161.318115 -26.5825653 ] solved after 0 constraint iterations from 300 voxels
amp2response: Shell b=7106: Response function [ 6709.48633 -3382.83374 1874.79749 -918.136597 379.606537 -120.274948 ] solved after 0 constraint iterations from 300 voxels
Command: amp2response dwi.mif gm_mask.mif dirs.mif gm.txt -shell 0,244,800,1536,4075,7106 -isotropic
Thank you for your time.
Best regards,
Achille