Hi @maxpietsch
Thanks for your reply.
I tried some code according to your reply. But they didn’t work.
I have b0 image (nodif.nii.gz), template image (MNI152_T1_1mm_brain.nii.gz), the tractography result (MNI152_T1_1mm_brain.nii.gz) and warp image got from FSL(MNI_2nodif_warp.nii.gz, nodif_2MNI_warp.nii.gz)
I want to transform the tck file to MNI space.
I tried three methods.
First, I used transformconvert and warpconvert to convert nodif_2MNI_warp.nii.gz to a warp format usable in MRtrix3, then used it to transform tck file to MNI space.
But, transformconvert allows to convert linear transformation matrices and warpconvert requires the warpfull file is 5D format. I don’t know to how to convert the warp I have got from FSL. Is there any other command available?
Second, I used the mrregister to get the warp then transform tck file to MNI space:
mrregister nodif.nii.gz MNI152_T1_1mm_brain.nii.gz -nl_warp_full warp_full.mif
warpconvert warp_full.mif warpfull2deformation w_t2s.mif -from 2 -template nodif.nii.gz
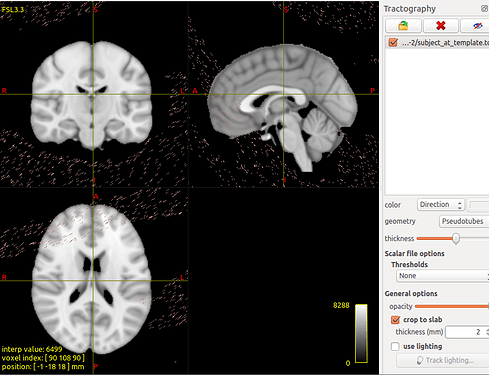
tcktransform SD_Stream_SIFT.tck w_t2s.mif subject_at_template.tck
But the result not correct.
The transformed tracts on the template:
Third, I want to try these code. But I only tried the command warpinit. Because I didn’t install ANTs and have never used it.
warpinit MNI152_T1_1mm_brain.nii.gz.nii.gz inv_identity_warp.nii.gz
inv_identity_warp.nii.gz seems to be right.
If I don’t use ANTs, how can I do that?
Stay healthy and stay happy!
Best regards,
Wu Xiao